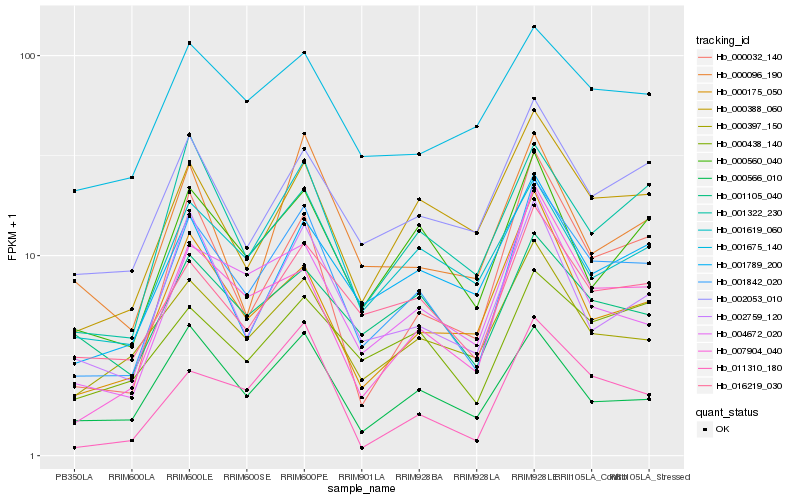
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001842_020 |
0.0 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 2 |
Hb_016219_030 |
0.1115888995 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_000560_040 |
0.1186634404 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007904_040 |
0.1206319536 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 5 |
Hb_002759_120 |
0.1220471144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002053_010 |
0.1227406991 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000388_060 |
0.1228951171 |
- |
- |
fructokinase [Manihot esculenta] |
| 8 |
Hb_004672_020 |
0.1241399379 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 9 |
Hb_000032_140 |
0.1241490904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000397_150 |
0.1255241357 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 11 |
Hb_000566_010 |
0.1289403391 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 12 |
Hb_000175_050 |
0.1312585379 |
- |
- |
- |
| 13 |
Hb_001789_200 |
0.1324240945 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001619_060 |
0.1326340667 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000438_140 |
0.1330548818 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 16 |
Hb_011310_180 |
0.1344093887 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001322_230 |
0.1352538428 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 18 |
Hb_001675_140 |
0.1433452918 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 19 |
Hb_000096_190 |
0.1443102252 |
- |
- |
unknown [Lotus japonicus] |
| 20 |
Hb_001105_040 |
0.1445201233 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |