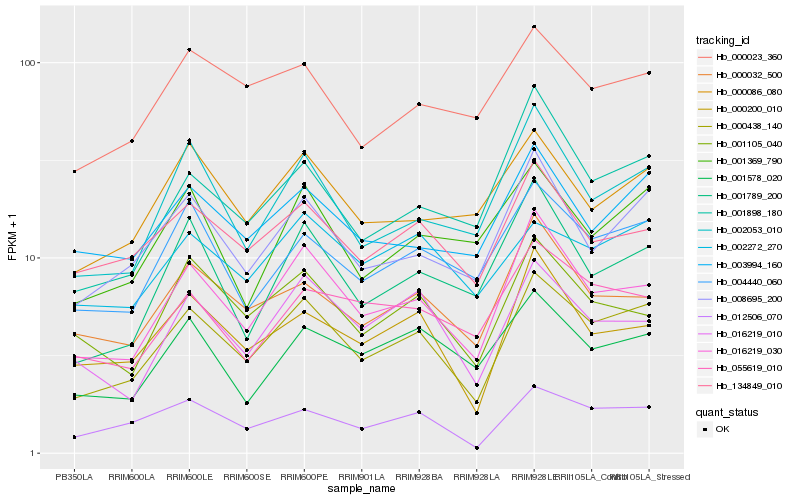
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_140 |
0.0 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 2 |
Hb_016219_030 |
0.0800174449 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_012506_070 |
0.0948807922 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 4 |
Hb_008695_200 |
0.098440404 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 5 |
Hb_004440_060 |
0.1084494761 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 6 |
Hb_002053_010 |
0.1087167534 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000200_010 |
0.1145306605 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 8 |
Hb_134849_010 |
0.1154237851 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 9 |
Hb_016219_010 |
0.1165501316 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 10 |
Hb_001898_180 |
0.118638771 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002272_270 |
0.119093418 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_000032_500 |
0.119328213 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 13 |
Hb_003994_160 |
0.1193991851 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 14 |
Hb_000086_080 |
0.1209003247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001789_200 |
0.1218597788 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001105_040 |
0.1219524135 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001369_790 |
0.1225830839 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000023_360 |
0.123022272 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001578_020 |
0.1248982579 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 20 |
Hb_055619_010 |
0.1256808765 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |