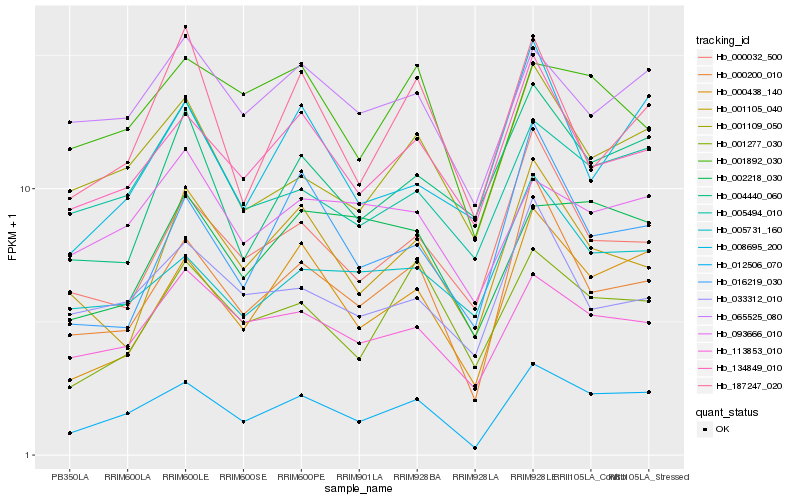
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012506_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 2 |
Hb_000438_140 |
0.0948807922 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 3 |
Hb_000200_010 |
0.1004725484 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 4 |
Hb_001109_050 |
0.1179603372 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 5 |
Hb_113853_010 |
0.119561391 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 6 |
Hb_134849_010 |
0.1216884852 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 7 |
Hb_001277_030 |
0.1253241704 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 8 |
Hb_005494_010 |
0.1297941536 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_000032_500 |
0.1329710344 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 10 |
Hb_065525_080 |
0.1356446222 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 11 |
Hb_002218_030 |
0.1391576461 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 12 |
Hb_004440_060 |
0.1399476896 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 13 |
Hb_016219_030 |
0.1408555667 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001892_030 |
0.1410646725 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase isoform X1 [Jatropha curcas] |
| 15 |
Hb_187247_020 |
0.1412286782 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 16 |
Hb_008695_200 |
0.1412732102 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 17 |
Hb_005731_160 |
0.1416648153 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_093666_010 |
0.1419325873 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 19 |
Hb_001105_040 |
0.1432005368 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_033312_010 |
0.1442285926 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |