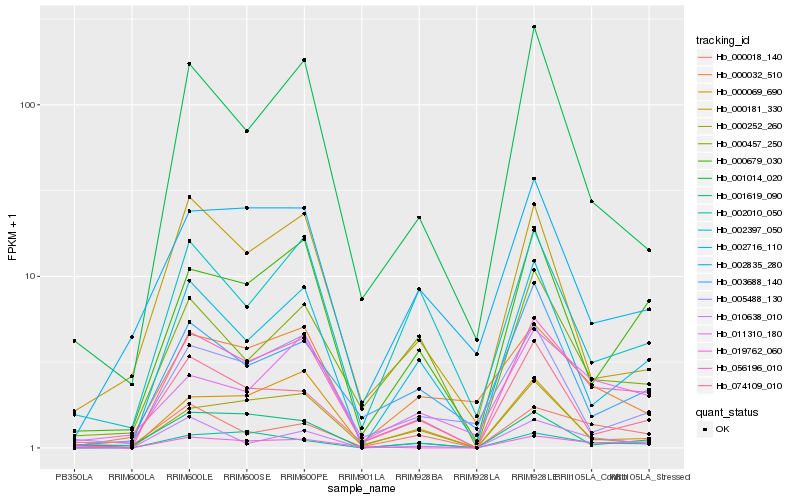
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056196_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 2 |
Hb_002716_110 |
0.1527195336 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 3 |
Hb_000679_030 |
0.1535525765 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 4 |
Hb_001014_020 |
0.166036824 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 5 |
Hb_074109_010 |
0.1754571202 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 6 |
Hb_019762_060 |
0.1826701972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000018_140 |
0.1833239803 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 8 |
Hb_005488_130 |
0.1900500028 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_011310_180 |
0.190873856 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_010638_010 |
0.1918039044 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 11 |
Hb_000069_690 |
0.1923201631 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 12 |
Hb_002010_050 |
0.1929657257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003688_140 |
0.1933962141 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 14 |
Hb_000032_510 |
0.1945288023 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_000457_250 |
0.1956674257 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 16 |
Hb_002397_050 |
0.1966043328 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000252_260 |
0.1974436314 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 18 |
Hb_002835_280 |
0.1996131152 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 19 |
Hb_001619_090 |
0.1998452827 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000181_330 |
0.2004400249 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |