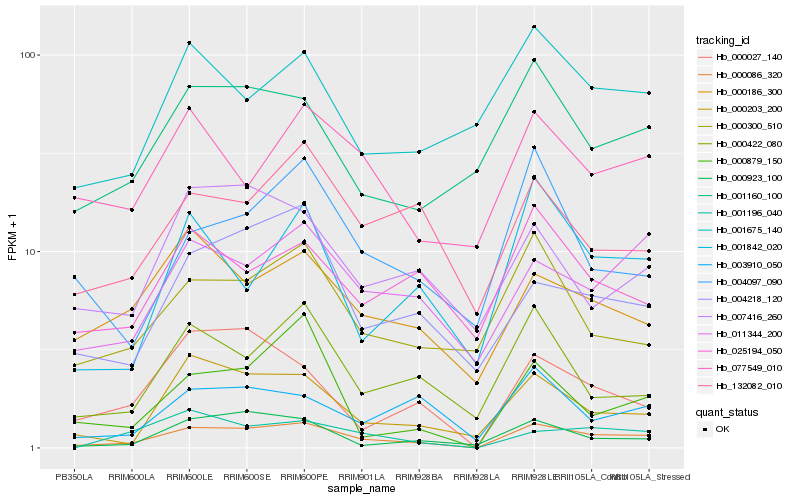
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 2 |
Hb_000203_200 |
0.1771158685 |
- |
- |
Pollen allergen Che a 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_011344_200 |
0.1850812092 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 4 |
Hb_004218_120 |
0.1861671735 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000422_080 |
0.1867684828 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_000300_510 |
0.1899545443 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 7 |
Hb_001842_020 |
0.1901746424 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 8 |
Hb_003910_050 |
0.1918721596 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 9 |
Hb_000027_140 |
0.1933232942 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 10 |
Hb_001160_100 |
0.1935473873 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000186_300 |
0.1947682935 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_132082_010 |
0.1994875829 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 13 |
Hb_000923_100 |
0.2000689103 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 14 |
Hb_004097_090 |
0.2018284808 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 15 |
Hb_001196_040 |
0.2025210713 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 16 |
Hb_077549_010 |
0.2032807965 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000879_150 |
0.2034392922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_025194_050 |
0.2039395709 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001675_140 |
0.2057029145 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 20 |
Hb_007416_260 |
0.2064147552 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |