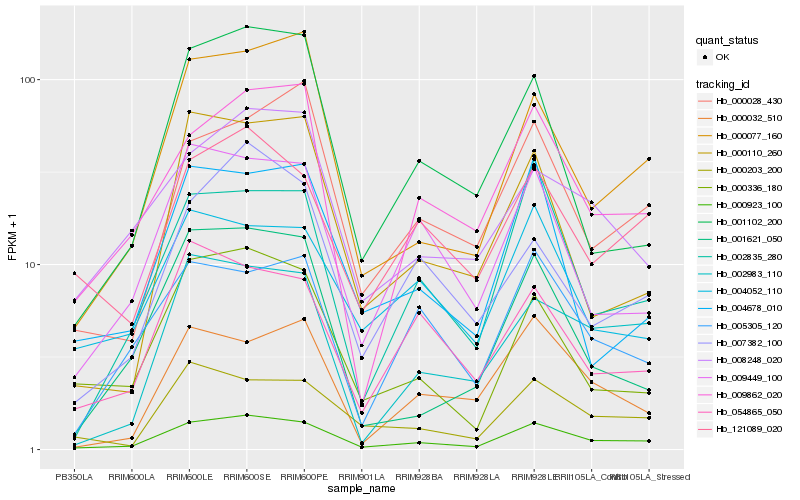
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000923_100 |
0.0 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 2 |
Hb_009862_020 |
0.1292324939 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 3 |
Hb_000077_160 |
0.1306295554 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 4 |
Hb_002835_280 |
0.1335697653 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 5 |
Hb_000336_180 |
0.1351536001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000203_200 |
0.1379201672 |
- |
- |
Pollen allergen Che a 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_121089_020 |
0.1421993225 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 8 |
Hb_009449_100 |
0.1431222551 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 9 |
Hb_000028_430 |
0.1457335676 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002983_110 |
0.1463426893 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 11 |
Hb_008248_020 |
0.1505067466 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001102_200 |
0.1510565704 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 13 |
Hb_004678_010 |
0.1530317197 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 14 |
Hb_005305_120 |
0.1540864865 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 15 |
Hb_054865_050 |
0.1541685974 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_001621_050 |
0.1557563985 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 17 |
Hb_007382_100 |
0.1577723222 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_004052_110 |
0.1585715718 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000110_260 |
0.1591900175 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 20 |
Hb_000032_510 |
0.1598310285 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |