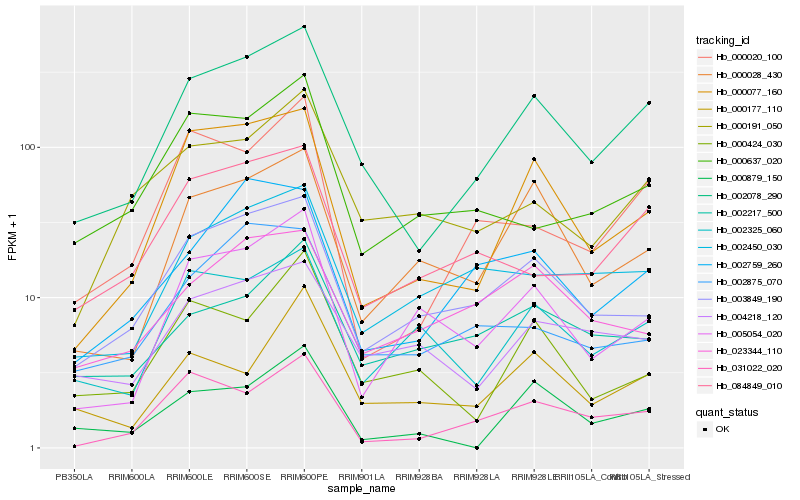
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 2 |
Hb_000077_160 |
0.1384697481 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 3 |
Hb_003849_190 |
0.1456530532 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 4 |
Hb_000028_430 |
0.1594758138 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031022_020 |
0.1610525219 |
- |
- |
- |
| 6 |
Hb_084849_010 |
0.1626285407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004218_120 |
0.1642990361 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002450_030 |
0.164630657 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 9 |
Hb_002759_260 |
0.1675143919 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 10 |
Hb_000191_050 |
0.1681117249 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 11 |
Hb_002325_060 |
0.1682640248 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 12 |
Hb_000020_100 |
0.1738974299 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 13 |
Hb_002217_500 |
0.1766483731 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 14 |
Hb_005054_020 |
0.1789544341 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 15 |
Hb_002875_070 |
0.1800455487 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 16 |
Hb_000879_150 |
0.1826307173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023344_110 |
0.1827064115 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 18 |
Hb_000177_110 |
0.1838732506 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 19 |
Hb_000424_030 |
0.186672569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000637_020 |
0.1868944826 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |