| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
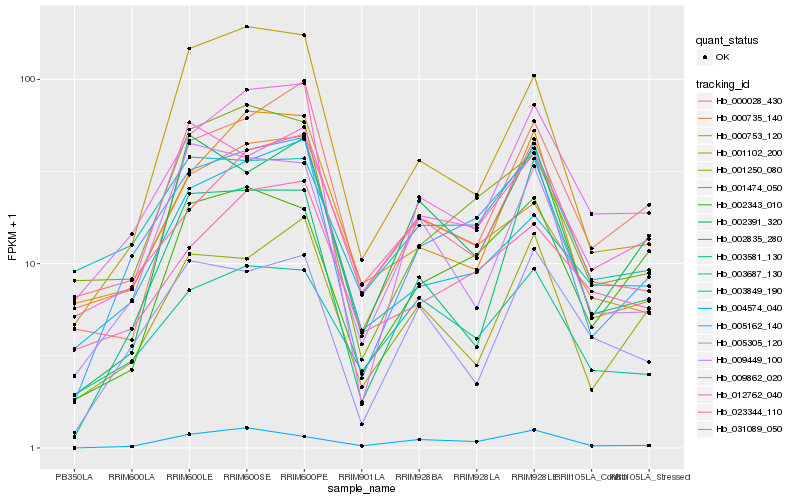
Hb_005162_140 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 2 |
Hb_009862_020 |
0.1346935634 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 3 |
Hb_001474_050 |
0.1352607229 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 4 |
Hb_012762_040 |
0.1540876838 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 5 |
Hb_001250_080 |
0.1546522349 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 6 |
Hb_002343_010 |
0.1548502691 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_003687_130 |
0.1565209536 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 8 |
Hb_000735_140 |
0.1578138213 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 9 |
Hb_003849_190 |
0.1599208548 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 10 |
Hb_000753_120 |
0.1627080631 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 11 |
Hb_001102_200 |
0.1636246918 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 12 |
Hb_003581_130 |
0.1656935744 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_009449_100 |
0.166445462 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 14 |
Hb_023344_110 |
0.1674919321 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 15 |
Hb_031089_050 |
0.1704143211 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_004574_040 |
0.1705878047 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 17 |
Hb_000028_430 |
0.1706100534 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002391_320 |
0.1708454268 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 19 |
Hb_002835_280 |
0.1708994617 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 20 |
Hb_005305_120 |
0.1716292935 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |