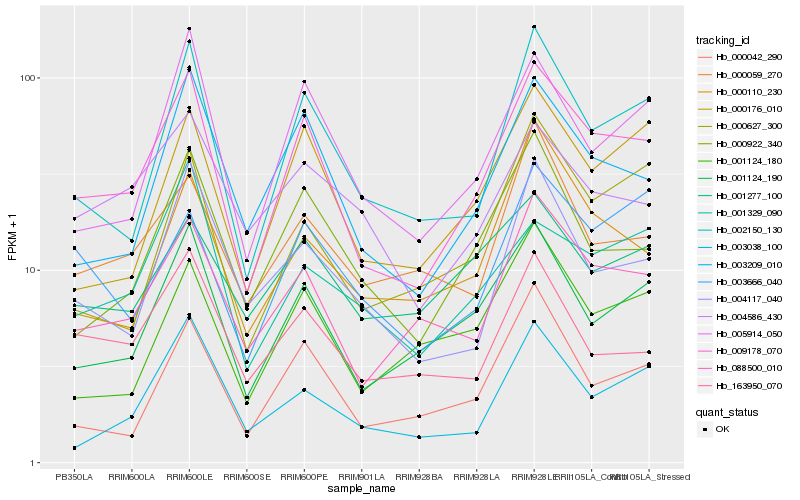
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009178_070 |
0.0 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002150_130 |
0.1152560337 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_088500_010 |
0.1156418265 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 4 |
Hb_001124_190 |
0.1180513369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003209_010 |
0.1199774509 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 6 |
Hb_005914_050 |
0.1277448458 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000922_340 |
0.1355663873 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 8 |
Hb_000176_010 |
0.1357536983 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000042_290 |
0.1419221974 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004117_040 |
0.1509339588 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 11 |
Hb_163950_070 |
0.1514027507 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 12 |
Hb_001124_180 |
0.1520280402 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 13 |
Hb_000110_230 |
0.1566305426 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000627_300 |
0.1573280981 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 15 |
Hb_003038_100 |
0.1592687151 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004586_430 |
0.1596024194 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000059_270 |
0.1599291311 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 18 |
Hb_001277_100 |
0.161769055 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_001329_090 |
0.1625083491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003666_040 |
0.1626347189 |
- |
- |
PREDICTED: uncharacterized protein LOC105639624 [Jatropha curcas] |