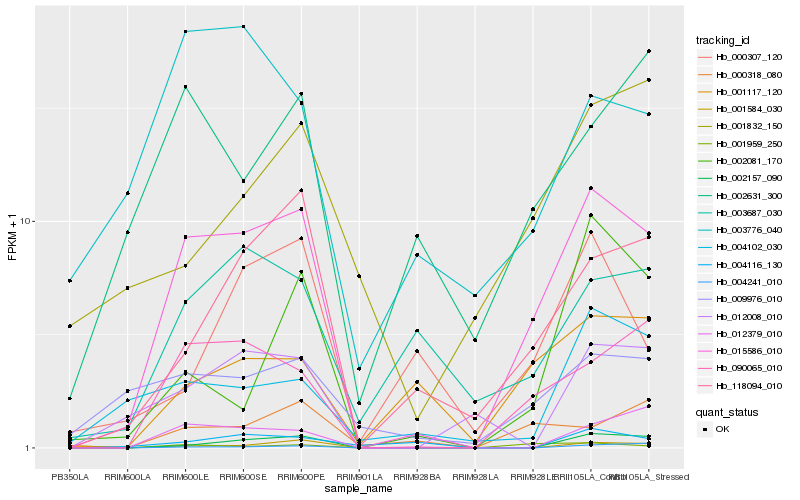
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015586_010 |
0.0 |
- |
- |
PREDICTED: flavonol sulfotransferase-like [Jatropha curcas] |
| 2 |
Hb_090065_010 |
0.1992590744 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 3 |
Hb_118094_010 |
0.2382716916 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 4 |
Hb_001584_030 |
0.2420894938 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Nelumbo nucifera] |
| 5 |
Hb_002157_090 |
0.2421224581 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 6 |
Hb_001959_250 |
0.2497190548 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 7 |
Hb_009976_010 |
0.2640924537 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 8 |
Hb_003687_030 |
0.264687766 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_001117_120 |
0.2660049786 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 10 |
Hb_012379_010 |
0.2674666222 |
- |
- |
- |
| 11 |
Hb_002631_300 |
0.2771150439 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 12 |
Hb_012008_010 |
0.2774419815 |
- |
- |
- |
| 13 |
Hb_004102_030 |
0.2806886551 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 14 |
Hb_000318_080 |
0.2843292155 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 15 |
Hb_004241_010 |
0.2902820625 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_000307_120 |
0.2930690716 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 17 |
Hb_003776_040 |
0.2952886999 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 18 |
Hb_002081_170 |
0.2953540182 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_004116_130 |
0.3047968056 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_001832_150 |
0.3055011315 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |