| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
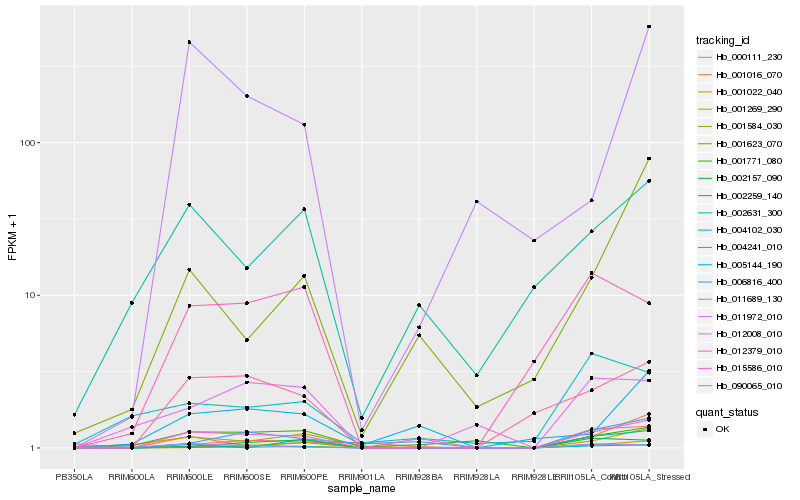
Hb_012379_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004241_010 |
0.167022449 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_090065_010 |
0.2090404134 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_002157_090 |
0.2518435423 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 5 |
Hb_001269_290 |
0.2585572566 |
- |
- |
PREDICTED: uncharacterized protein LOC103438859 [Malus domestica] |
| 6 |
Hb_015586_010 |
0.2674666222 |
- |
- |
PREDICTED: flavonol sulfotransferase-like [Jatropha curcas] |
| 7 |
Hb_000111_230 |
0.2758600338 |
- |
- |
- |
| 8 |
Hb_012008_010 |
0.2764799077 |
- |
- |
- |
| 9 |
Hb_001584_030 |
0.2807981775 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Nelumbo nucifera] |
| 10 |
Hb_011972_010 |
0.2884567951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001771_080 |
0.2982917353 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 12 |
Hb_006816_400 |
0.2989164383 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 13 |
Hb_001022_040 |
0.3013722669 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 14 |
Hb_002631_300 |
0.3057890918 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 15 |
Hb_011689_130 |
0.3085935459 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001016_070 |
0.3107734018 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 17 |
Hb_005144_190 |
0.3158257704 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 18 |
Hb_004102_030 |
0.3189539121 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 19 |
Hb_002259_140 |
0.3223792931 |
- |
- |
- |
| 20 |
Hb_001623_070 |
0.3271643616 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |