| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
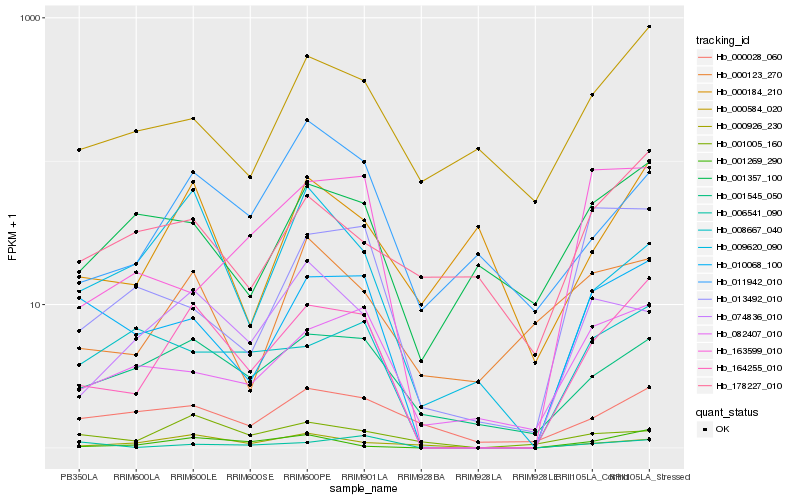
Hb_164255_010 |
0.0 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 2 |
Hb_010068_100 |
0.2155964109 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001269_290 |
0.2175136093 |
- |
- |
PREDICTED: uncharacterized protein LOC103438859 [Malus domestica] |
| 4 |
Hb_074836_010 |
0.2294400425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001545_050 |
0.2305330688 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 6 |
Hb_082407_010 |
0.2490481315 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_000926_230 |
0.2539623231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008667_040 |
0.2543834607 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 9 |
Hb_000584_020 |
0.2633280538 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 10 |
Hb_006541_090 |
0.2647193957 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 11 |
Hb_000028_060 |
0.2658697999 |
- |
- |
- |
| 12 |
Hb_009620_090 |
0.26714981 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 13 |
Hb_001357_100 |
0.2703356434 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 14 |
Hb_000184_210 |
0.2707782624 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 15 |
Hb_001005_160 |
0.2730517484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011942_010 |
0.2736595991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_163599_010 |
0.2751173464 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 18 |
Hb_000123_270 |
0.2789680234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_178227_010 |
0.2799882 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 20 |
Hb_013492_010 |
0.2805546186 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |