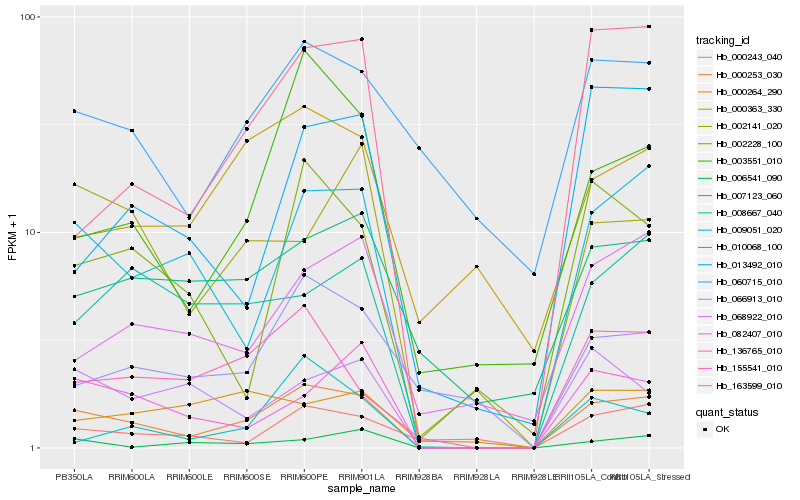
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000253_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 2 |
Hb_010068_100 |
0.1666538916 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_163599_010 |
0.199362676 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 4 |
Hb_136765_010 |
0.2035325607 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_155541_010 |
0.2050204036 |
- |
- |
- |
| 6 |
Hb_003551_010 |
0.2109017973 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 7 |
Hb_008667_040 |
0.2119574121 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 8 |
Hb_002228_100 |
0.2198177117 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_002141_020 |
0.2223638473 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 10 |
Hb_068922_010 |
0.2237222873 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 11 |
Hb_007123_060 |
0.2238928266 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 12 |
Hb_082407_010 |
0.2242188445 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_060715_010 |
0.2318935859 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 14 |
Hb_006541_090 |
0.2332324745 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 15 |
Hb_000243_040 |
0.2340864862 |
- |
- |
- |
| 16 |
Hb_000363_330 |
0.2382990107 |
- |
- |
- |
| 17 |
Hb_009051_020 |
0.2401658831 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 18 |
Hb_066913_010 |
0.2407955529 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 19 |
Hb_013492_010 |
0.2423250401 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 20 |
Hb_000264_290 |
0.2430859418 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |