| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
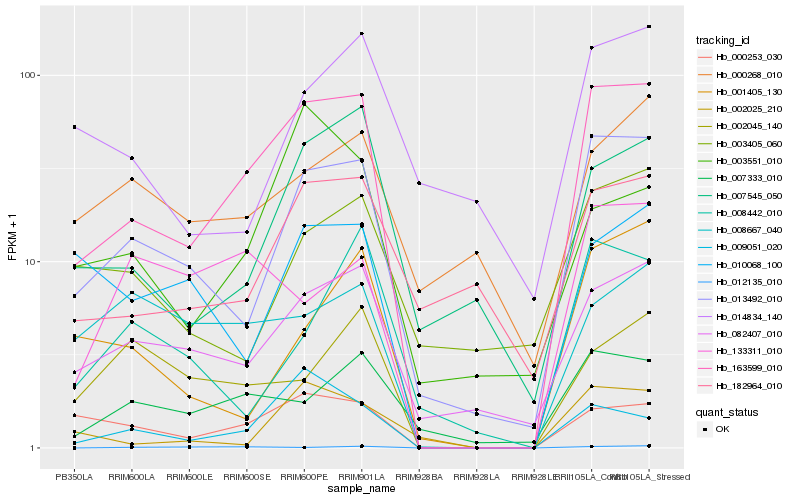
Hb_163599_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 2 |
Hb_013492_010 |
0.1527248524 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 3 |
Hb_082407_010 |
0.1890966708 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_000253_030 |
0.199362676 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 5 |
Hb_007333_010 |
0.2150700704 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_007545_050 |
0.2152859085 |
- |
- |
- |
| 7 |
Hb_009051_020 |
0.2311107339 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 8 |
Hb_182964_010 |
0.2334183881 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002025_210 |
0.2382203723 |
- |
- |
- |
| 10 |
Hb_001405_130 |
0.2391197465 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 11 |
Hb_010068_100 |
0.2405608337 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 12 |
Hb_133311_010 |
0.2526284323 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 13 |
Hb_008667_040 |
0.2561974509 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 14 |
Hb_002045_140 |
0.2565570828 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 15 |
Hb_000268_010 |
0.2573219203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003405_060 |
0.258132548 |
- |
- |
- |
| 17 |
Hb_008442_010 |
0.259069583 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 18 |
Hb_014834_140 |
0.2594038089 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 19 |
Hb_012135_010 |
0.2622073212 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 20 |
Hb_003551_010 |
0.2633839769 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |