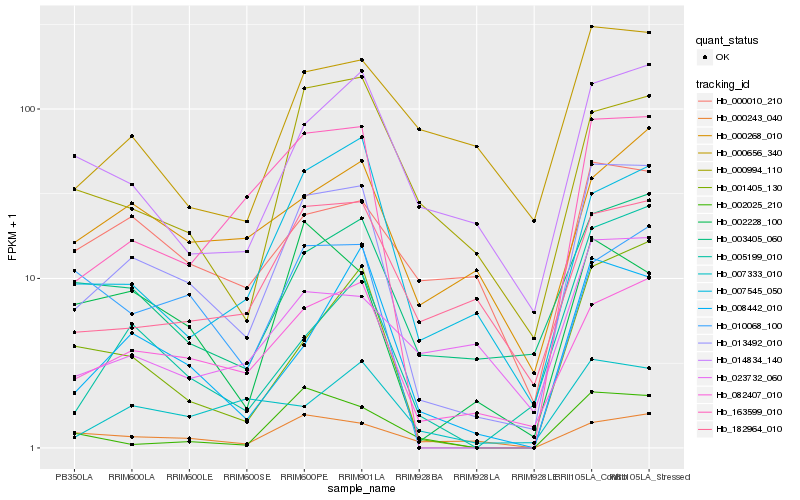
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013492_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 2 |
Hb_082407_010 |
0.1458023549 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_163599_010 |
0.1527248524 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 4 |
Hb_003405_060 |
0.1751475319 |
- |
- |
- |
| 5 |
Hb_001405_130 |
0.1798886281 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 6 |
Hb_008442_010 |
0.1811637195 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 7 |
Hb_014834_140 |
0.195874713 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 8 |
Hb_007545_050 |
0.2060315779 |
- |
- |
- |
| 9 |
Hb_010068_100 |
0.2065817968 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002228_100 |
0.2081354702 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_000994_110 |
0.208422082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000243_040 |
0.2088059512 |
- |
- |
- |
| 13 |
Hb_002025_210 |
0.2096677693 |
- |
- |
- |
| 14 |
Hb_182964_010 |
0.211057366 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007333_010 |
0.2117171573 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000010_210 |
0.2160086869 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 17 |
Hb_000268_010 |
0.2161986291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005199_010 |
0.2174435043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000656_340 |
0.2200161825 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 20 |
Hb_023732_060 |
0.2206934526 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |