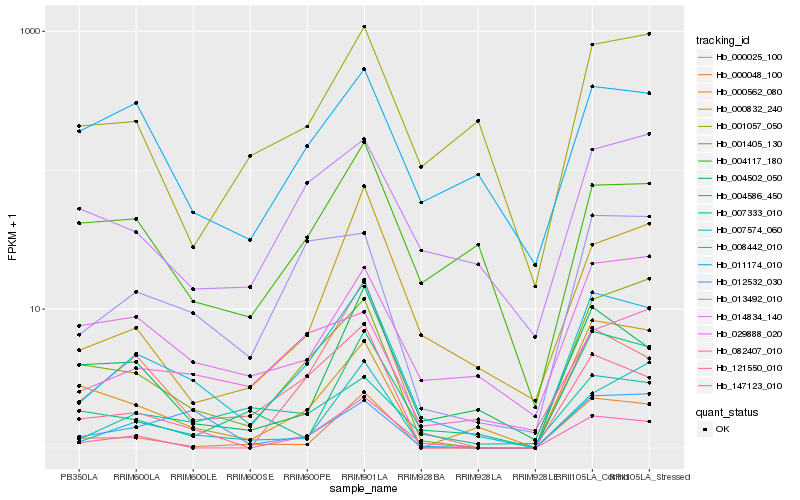
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008442_010 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 2 |
Hb_013492_010 |
0.1811637195 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 3 |
Hb_000025_100 |
0.1876583141 |
- |
- |
- |
| 4 |
Hb_001405_130 |
0.1884824154 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 5 |
Hb_029888_020 |
0.1906697382 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 6 |
Hb_121550_010 |
0.1947535315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_147123_010 |
0.2000756103 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_007333_010 |
0.2010876802 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_012532_030 |
0.2029623782 |
- |
- |
- |
| 10 |
Hb_007574_060 |
0.2077041275 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |
| 11 |
Hb_004502_050 |
0.2081503971 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 12 |
Hb_000048_100 |
0.209830237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000562_080 |
0.2101423547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_014834_140 |
0.2126096255 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 15 |
Hb_082407_010 |
0.2144391962 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_000832_240 |
0.2169336699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004586_450 |
0.2185676271 |
- |
- |
- |
| 18 |
Hb_001057_050 |
0.2200456932 |
- |
- |
hypothetical protein POPTR_0003s15960g, partial [Populus trichocarpa] |
| 19 |
Hb_004117_180 |
0.2206536782 |
- |
- |
- |
| 20 |
Hb_011174_010 |
0.2228414006 |
- |
- |
hypothetical protein JCGZ_10122 [Jatropha curcas] |