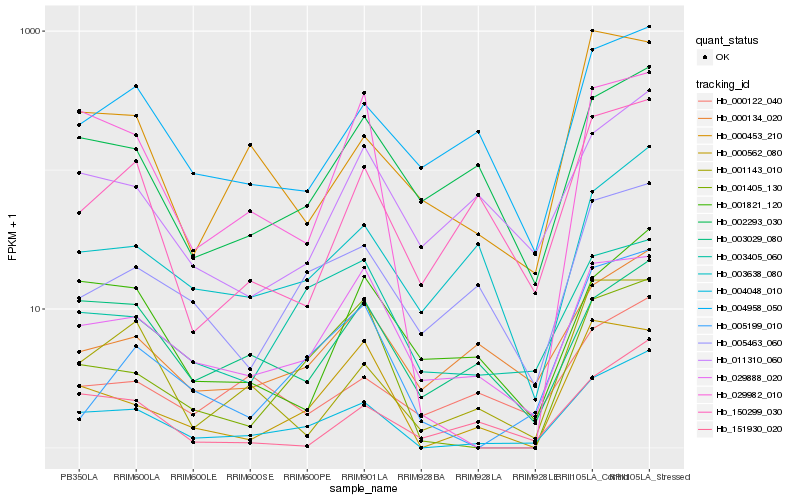
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004048_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 2 |
Hb_001405_130 |
0.188432471 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 3 |
Hb_029982_010 |
0.193237194 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 4 |
Hb_000134_020 |
0.1947046128 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 5 |
Hb_003638_080 |
0.1991910651 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 6 |
Hb_005199_010 |
0.2004233791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_029888_020 |
0.2009067036 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 8 |
Hb_004958_050 |
0.2022603743 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 9 |
Hb_002293_030 |
0.2036874309 |
- |
- |
eukaryotic translation initiation factor 5A isoform VI [Hevea brasiliensis] |
| 10 |
Hb_005463_060 |
0.2041045951 |
- |
- |
PREDICTED: uncharacterized protein LOC105632515 [Jatropha curcas] |
| 11 |
Hb_011310_060 |
0.2067880502 |
- |
- |
PREDICTED: uncharacterized protein At2g34160-like [Jatropha curcas] |
| 12 |
Hb_151930_020 |
0.2079920605 |
- |
- |
Wound-induced protein WIN1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001143_010 |
0.2081035464 |
- |
- |
- |
| 14 |
Hb_000562_080 |
0.2090762089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_150299_030 |
0.2127543517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003029_080 |
0.2138623552 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 17 |
Hb_000122_040 |
0.2143874586 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 18 |
Hb_000453_210 |
0.2158246275 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 19 |
Hb_003405_060 |
0.2171093447 |
- |
- |
- |
| 20 |
Hb_001821_120 |
0.2178429079 |
- |
- |
PREDICTED: uncharacterized protein LOC105643956 [Jatropha curcas] |