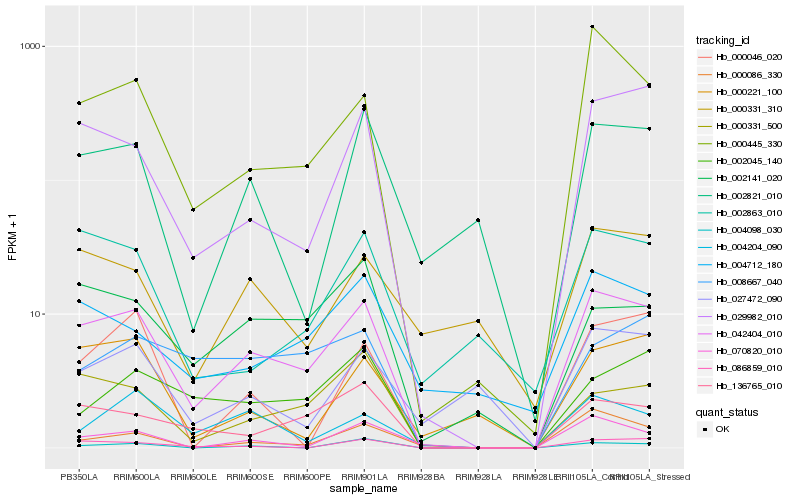
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042404_010 |
0.0 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 2 |
Hb_000445_330 |
0.1658782949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_029982_010 |
0.1714677195 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 4 |
Hb_136765_010 |
0.1887762753 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_086859_010 |
0.1954173926 |
- |
- |
- |
| 6 |
Hb_000331_500 |
0.1980012996 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_070820_010 |
0.2033172354 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 8 |
Hb_002141_020 |
0.2082942969 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 9 |
Hb_004712_180 |
0.2091225149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000086_330 |
0.216329445 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_002821_010 |
0.2167663344 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 12 |
Hb_002863_010 |
0.2191570879 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 13 |
Hb_002045_140 |
0.2233762817 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 14 |
Hb_004204_090 |
0.2238430879 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 15 |
Hb_008667_040 |
0.2268151037 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 16 |
Hb_000331_310 |
0.2278992011 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 17 |
Hb_027472_090 |
0.2279003267 |
- |
- |
- |
| 18 |
Hb_000221_100 |
0.228805641 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |
| 19 |
Hb_004098_030 |
0.2290429782 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 20 |
Hb_000046_020 |
0.2294705766 |
- |
- |
- |