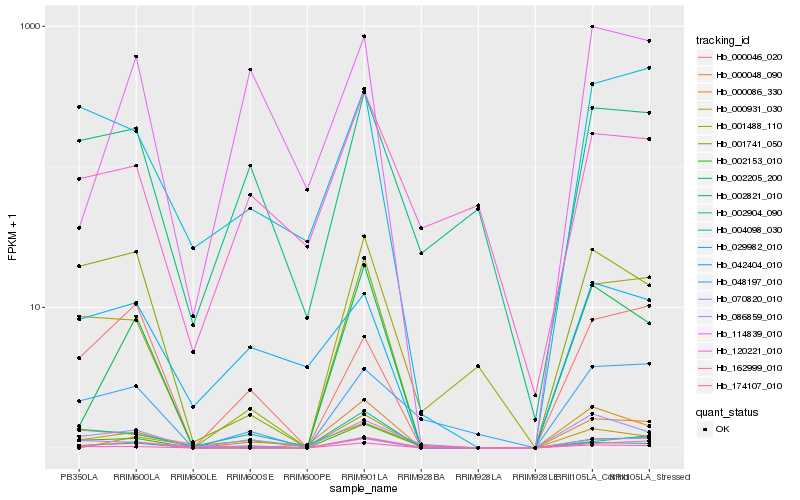
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004098_030 |
0.0 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 2 |
Hb_070820_010 |
0.1930228096 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 3 |
Hb_002821_010 |
0.2036038863 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 4 |
Hb_162999_010 |
0.2096877068 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_086859_010 |
0.2224036796 |
- |
- |
- |
| 6 |
Hb_001488_110 |
0.2254911746 |
- |
- |
- |
| 7 |
Hb_042404_010 |
0.2290429782 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 8 |
Hb_002153_010 |
0.2308243389 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 9 |
Hb_000046_020 |
0.2319052002 |
- |
- |
- |
| 10 |
Hb_001741_050 |
0.2357574888 |
- |
- |
- |
| 11 |
Hb_174107_010 |
0.2366147944 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 12 |
Hb_114839_010 |
0.2412868868 |
- |
- |
- |
| 13 |
Hb_048197_010 |
0.2520526592 |
- |
- |
- |
| 14 |
Hb_002904_090 |
0.2531705925 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420-like [Vitis vinifera] |
| 15 |
Hb_120221_010 |
0.2537065834 |
- |
- |
- |
| 16 |
Hb_000086_330 |
0.2546040111 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_029982_010 |
0.2554810391 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 18 |
Hb_000048_090 |
0.2576273969 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 19 |
Hb_000931_030 |
0.258211214 |
- |
- |
PREDICTED: uncharacterized protein LOC105130278 [Populus euphratica] |
| 20 |
Hb_002205_200 |
0.2588304124 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |