| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
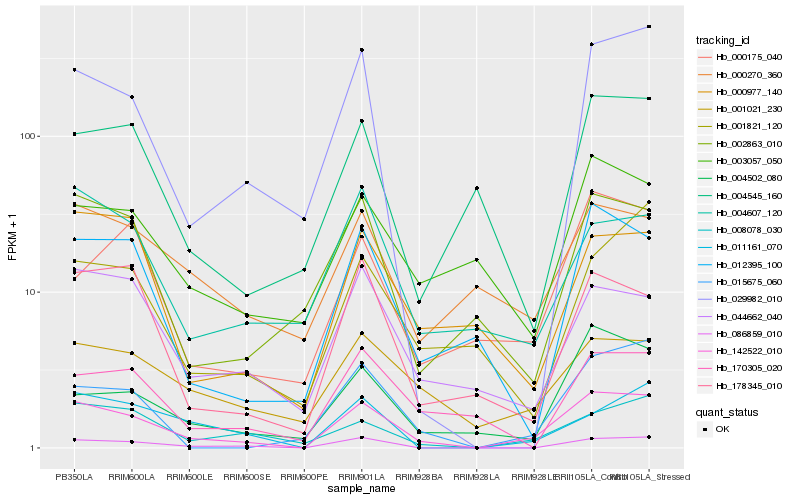
Hb_142522_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_086859_010 |
0.1754679713 |
- |
- |
- |
| 3 |
Hb_008078_030 |
0.1894271685 |
- |
- |
- |
| 4 |
Hb_029982_010 |
0.1968524978 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 5 |
Hb_001021_230 |
0.1975724664 |
- |
- |
- |
| 6 |
Hb_015675_060 |
0.200307957 |
- |
- |
- |
| 7 |
Hb_004502_080 |
0.2010342533 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 8 |
Hb_044662_040 |
0.2043912152 |
- |
- |
- |
| 9 |
Hb_012395_100 |
0.2045081182 |
- |
- |
eukaryotic translation initiation factor 4e, putative [Ricinus communis] |
| 10 |
Hb_178345_010 |
0.2078657745 |
- |
- |
hypothetical protein PHAVU_004G084200g [Phaseolus vulgaris] |
| 11 |
Hb_011161_070 |
0.2103743335 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X1 [Populus euphratica] |
| 12 |
Hb_002863_010 |
0.2117044836 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 13 |
Hb_000175_040 |
0.2129421974 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 14 |
Hb_004607_120 |
0.2175934743 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 8 [Jatropha curcas] |
| 15 |
Hb_003057_050 |
0.2185038714 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 16 |
Hb_000977_140 |
0.2199577462 |
- |
- |
PREDICTED: uncharacterized protein LOC105801335 [Gossypium raimondii] |
| 17 |
Hb_001821_120 |
0.2218857933 |
- |
- |
PREDICTED: uncharacterized protein LOC105643956 [Jatropha curcas] |
| 18 |
Hb_000270_360 |
0.2219585077 |
- |
- |
PREDICTED: serine/threonine-protein kinase pakF-like [Jatropha curcas] |
| 19 |
Hb_170305_020 |
0.2228821816 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 20 |
Hb_004545_160 |
0.2235773669 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |