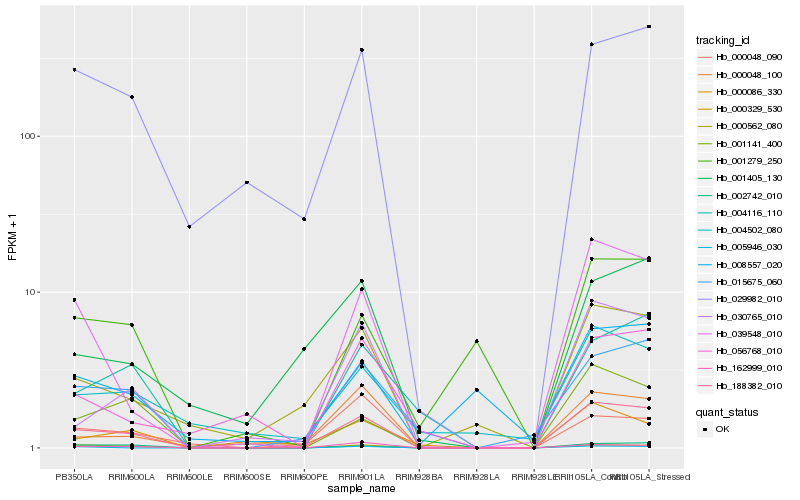
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_188382_010 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_015675_060 |
0.1705853314 |
- |
- |
- |
| 3 |
Hb_039548_010 |
0.1706688259 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 4 |
Hb_030765_010 |
0.1803118063 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 5 |
Hb_002742_010 |
0.1832416977 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 6 |
Hb_029982_010 |
0.1851006334 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 7 |
Hb_000562_080 |
0.1854544663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001141_400 |
0.196604501 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 9 |
Hb_162999_010 |
0.1987656952 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_000048_100 |
0.1996681171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000329_530 |
0.2009484177 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 12 |
Hb_004116_110 |
0.2083816276 |
- |
- |
hypothetical protein MTR_3g064100 [Medicago truncatula] |
| 13 |
Hb_000086_330 |
0.2217077917 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_004502_080 |
0.2241851971 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 15 |
Hb_000048_090 |
0.2274485614 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 16 |
Hb_001405_130 |
0.2278790807 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 17 |
Hb_001279_250 |
0.2361328434 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 18 |
Hb_008557_020 |
0.2368653364 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 19 |
Hb_005946_030 |
0.2372853085 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 20 |
Hb_056768_010 |
0.2378471142 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Cicer arietinum] |