| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
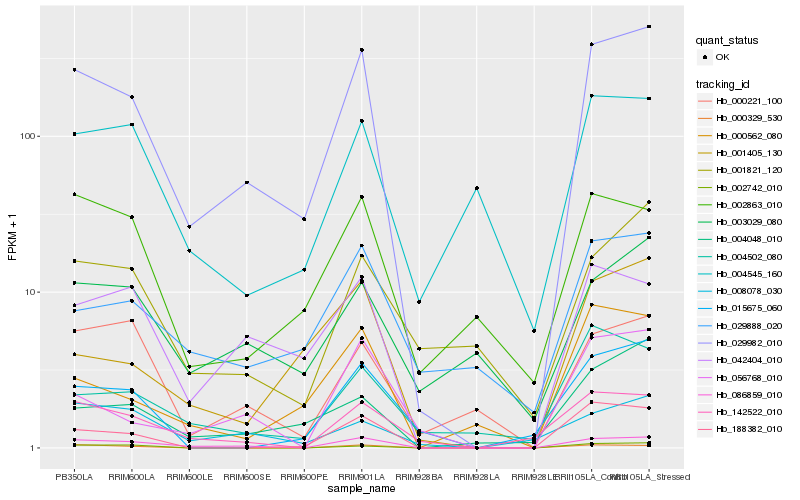
Hb_029982_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 2 |
Hb_086859_010 |
0.1233308021 |
- |
- |
- |
| 3 |
Hb_042404_010 |
0.1714677195 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 4 |
Hb_188382_010 |
0.1851006334 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_004048_010 |
0.193237194 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 6 |
Hb_001405_130 |
0.1953241471 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 7 |
Hb_142522_010 |
0.1968524978 |
- |
- |
- |
| 8 |
Hb_000562_080 |
0.2019101033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_015675_060 |
0.2026660884 |
- |
- |
- |
| 10 |
Hb_029888_020 |
0.2081984773 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 11 |
Hb_000221_100 |
0.2082666509 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |
| 12 |
Hb_056768_010 |
0.2104647813 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Cicer arietinum] |
| 13 |
Hb_002863_010 |
0.2105504256 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 14 |
Hb_001821_120 |
0.2128340314 |
- |
- |
PREDICTED: uncharacterized protein LOC105643956 [Jatropha curcas] |
| 15 |
Hb_002742_010 |
0.213917756 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 16 |
Hb_003029_080 |
0.2149551082 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 17 |
Hb_004502_080 |
0.2159219487 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 18 |
Hb_000329_530 |
0.2166843292 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 19 |
Hb_004545_160 |
0.2181517628 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 20 |
Hb_008078_030 |
0.2187690244 |
- |
- |
- |