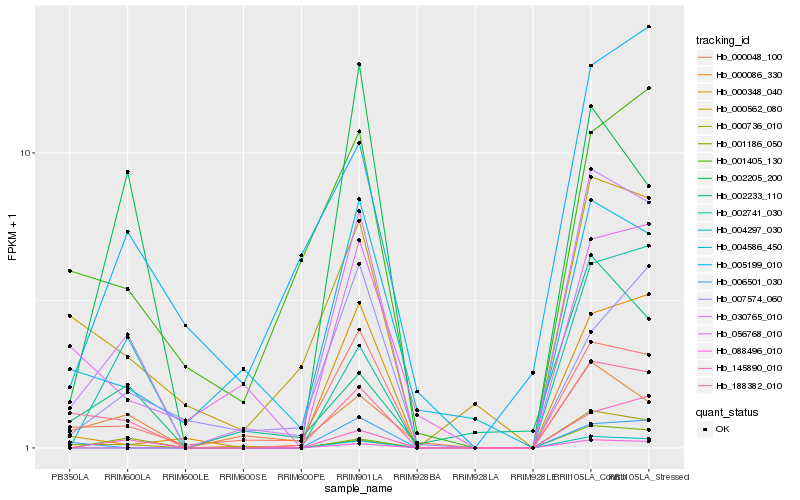
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030765_010 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 2 |
Hb_000048_100 |
0.1397175677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_188382_010 |
0.1803118063 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_001186_050 |
0.200572867 |
- |
- |
- |
| 5 |
Hb_000086_330 |
0.2119707724 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_000348_040 |
0.2146354802 |
- |
- |
- |
| 7 |
Hb_000736_010 |
0.2148730786 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 8 |
Hb_004586_450 |
0.2164935423 |
- |
- |
- |
| 9 |
Hb_000562_080 |
0.2201475294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_145890_010 |
0.2209752057 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002741_030 |
0.2214068706 |
- |
- |
- |
| 12 |
Hb_004297_030 |
0.2265056637 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 13 |
Hb_002233_110 |
0.2296044832 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 14 |
Hb_002205_200 |
0.2312833399 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 15 |
Hb_088496_010 |
0.2314146138 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_056768_010 |
0.2369795895 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Cicer arietinum] |
| 17 |
Hb_007574_060 |
0.2425894038 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |
| 18 |
Hb_001405_130 |
0.2475785248 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 19 |
Hb_006501_030 |
0.2477592592 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 20 |
Hb_005199_010 |
0.2487900846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |