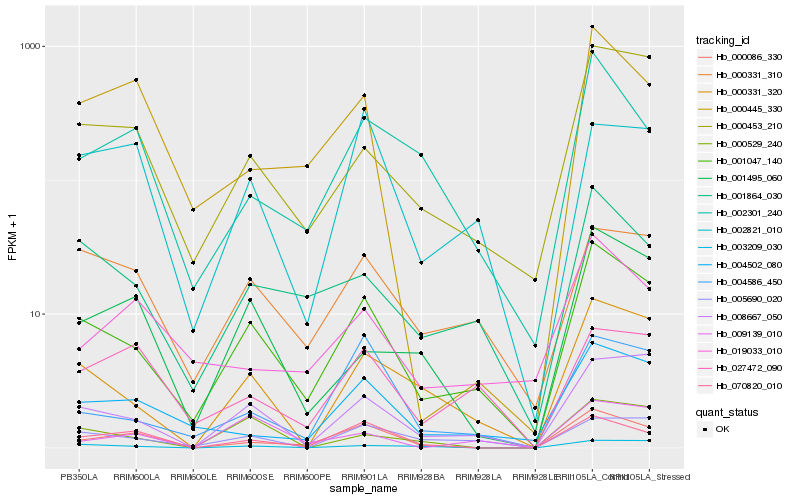
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001047_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_000331_320 |
0.1587862202 |
- |
- |
- |
| 3 |
Hb_001864_030 |
0.1619088591 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 4 |
Hb_008667_050 |
0.1635759488 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 5 |
Hb_000453_210 |
0.1769370791 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 6 |
Hb_000331_310 |
0.1880414777 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 7 |
Hb_004502_080 |
0.1907995305 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 8 |
Hb_005690_020 |
0.1928696824 |
- |
- |
hypothetical protein POPTR_0006s08200g [Populus trichocarpa] |
| 9 |
Hb_000086_330 |
0.1945538519 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_004586_450 |
0.2073382415 |
- |
- |
- |
| 11 |
Hb_002301_240 |
0.2086329924 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 12 |
Hb_003209_030 |
0.2092192935 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 13 |
Hb_001495_060 |
0.2145323891 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 14 |
Hb_070820_010 |
0.2210832992 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 15 |
Hb_000529_240 |
0.2244299984 |
- |
- |
- |
| 16 |
Hb_000445_330 |
0.2272979986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_027472_090 |
0.228900292 |
- |
- |
- |
| 18 |
Hb_019033_010 |
0.2296800389 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 19 |
Hb_002821_010 |
0.2304120487 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 20 |
Hb_009139_010 |
0.2324622179 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |