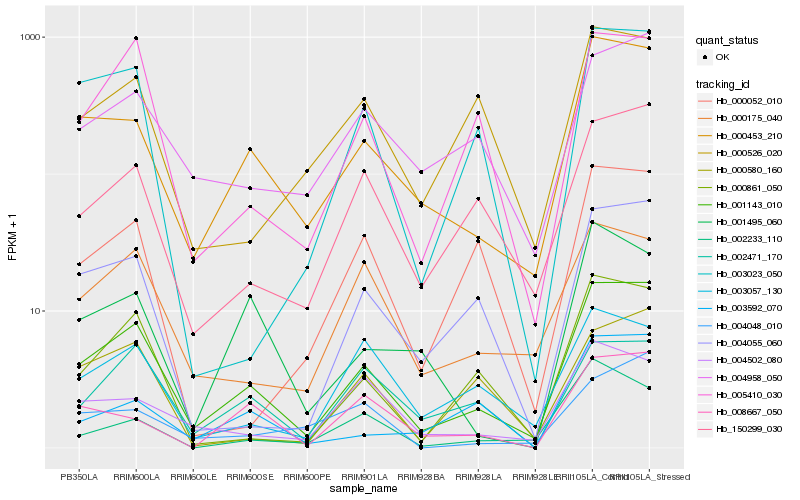
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000453_210 |
0.1244241929 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 3 |
Hb_000861_050 |
0.1635565002 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 4 |
Hb_150299_030 |
0.1644433588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005410_030 |
0.1708176699 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 6 |
Hb_004055_060 |
0.1725839143 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 7 |
Hb_004958_050 |
0.1800151976 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 8 |
Hb_003023_050 |
0.1873273076 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 9 |
Hb_000175_040 |
0.1878848967 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 10 |
Hb_000580_160 |
0.1974937621 |
- |
- |
Nitrate transporter 1.5 [Morus notabilis] |
| 11 |
Hb_002471_170 |
0.19845301 |
- |
- |
- |
| 12 |
Hb_004502_080 |
0.198795576 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 13 |
Hb_000526_020 |
0.201706999 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 14 |
Hb_000052_010 |
0.2019847594 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 15 |
Hb_003057_130 |
0.2028572019 |
- |
- |
- |
| 16 |
Hb_001495_060 |
0.2037751318 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 17 |
Hb_008667_050 |
0.2038519561 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 18 |
Hb_003592_070 |
0.2042365371 |
- |
- |
- |
| 19 |
Hb_002233_110 |
0.2066901257 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 20 |
Hb_004048_010 |
0.2081035464 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |