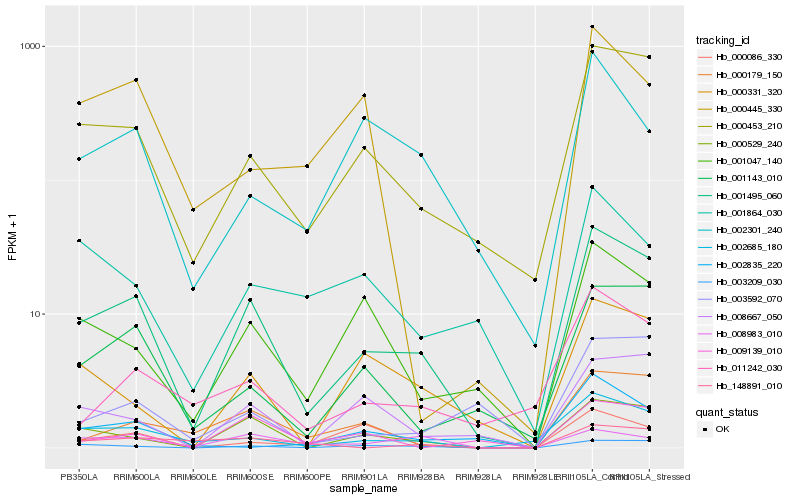
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001495_060 |
0.0 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 2 |
Hb_000453_210 |
0.1540596622 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 3 |
Hb_000529_240 |
0.1775299213 |
- |
- |
- |
| 4 |
Hb_002685_180 |
0.1882155102 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003209_030 |
0.1942177101 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 6 |
Hb_001143_010 |
0.2037751318 |
- |
- |
- |
| 7 |
Hb_001047_140 |
0.2145323891 |
- |
- |
- |
| 8 |
Hb_008667_050 |
0.2194152289 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 9 |
Hb_000331_320 |
0.223250428 |
- |
- |
- |
| 10 |
Hb_011242_030 |
0.2233439501 |
- |
- |
- |
| 11 |
Hb_000179_150 |
0.2242601827 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 12 |
Hb_002301_240 |
0.2484339028 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 13 |
Hb_009139_010 |
0.2484663276 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |
| 14 |
Hb_001864_030 |
0.2493081554 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 15 |
Hb_003592_070 |
0.2514123427 |
- |
- |
- |
| 16 |
Hb_000086_330 |
0.2589234664 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_002835_220 |
0.2593913512 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 18 |
Hb_148891_010 |
0.2630478815 |
- |
- |
- |
| 19 |
Hb_000445_330 |
0.2669522106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008983_010 |
0.2673476243 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |