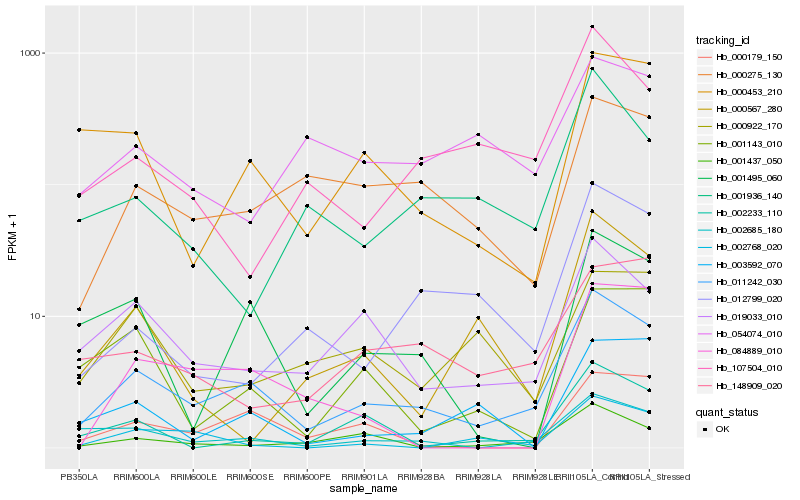
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011242_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_002685_180 |
0.1909087929 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_019033_010 |
0.1954930634 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 4 |
Hb_000453_210 |
0.2153594393 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 5 |
Hb_001437_050 |
0.218981298 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 6 |
Hb_000275_130 |
0.2212174368 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |
| 7 |
Hb_001495_060 |
0.2233439501 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 8 |
Hb_000179_150 |
0.2261059472 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 9 |
Hb_002233_110 |
0.2289019798 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 10 |
Hb_107504_010 |
0.231157129 |
- |
- |
- |
| 11 |
Hb_148909_020 |
0.2354140141 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_001936_140 |
0.2369937497 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 1 [Jatropha curcas] |
| 13 |
Hb_012799_020 |
0.2387967562 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_001143_010 |
0.2464534512 |
- |
- |
- |
| 15 |
Hb_003592_070 |
0.2476444793 |
- |
- |
- |
| 16 |
Hb_002768_020 |
0.2482052687 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 17 |
Hb_000567_280 |
0.2513186138 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 18 |
Hb_000922_170 |
0.2535663149 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 19 |
Hb_054074_010 |
0.253822791 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 20 |
Hb_084889_010 |
0.2601670562 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |