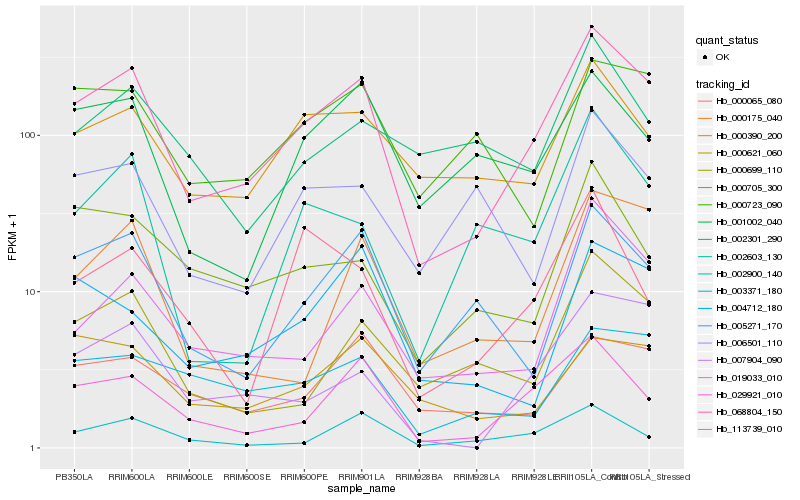
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_029921_010 |
0.1522143399 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 3 |
Hb_000390_200 |
0.1693733216 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 4 |
Hb_005271_170 |
0.1703481381 |
- |
- |
PREDICTED: tetraspanin-10 isoform X3 [Vitis vinifera] |
| 5 |
Hb_019033_010 |
0.1754451431 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 6 |
Hb_000699_110 |
0.1784648629 |
- |
- |
hypothetical protein POPTR_0013s04180g [Populus trichocarpa] |
| 7 |
Hb_000065_080 |
0.1820230261 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, chloroplastic-like [Nelumbo nucifera] |
| 8 |
Hb_007904_090 |
0.1833495325 |
- |
- |
PREDICTED: uncharacterized protein LOC100261196 [Vitis vinifera] |
| 9 |
Hb_003371_180 |
0.1842210256 |
- |
- |
- |
| 10 |
Hb_001002_040 |
0.1864067276 |
- |
- |
PREDICTED: ATP-citrate synthase beta chain protein 2 [Gossypium raimondii] |
| 11 |
Hb_002900_140 |
0.1882536222 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_000175_040 |
0.1895631184 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 13 |
Hb_000705_300 |
0.1905024337 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 14 |
Hb_002603_130 |
0.1925522429 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 15 |
Hb_000723_090 |
0.1946133425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000621_060 |
0.1946986333 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 17 |
Hb_004712_180 |
0.1956753216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006501_110 |
0.2015275563 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 19 |
Hb_113739_010 |
0.204116063 |
- |
- |
hypothetical protein JCGZ_06379 [Jatropha curcas] |
| 20 |
Hb_002301_290 |
0.208197043 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |