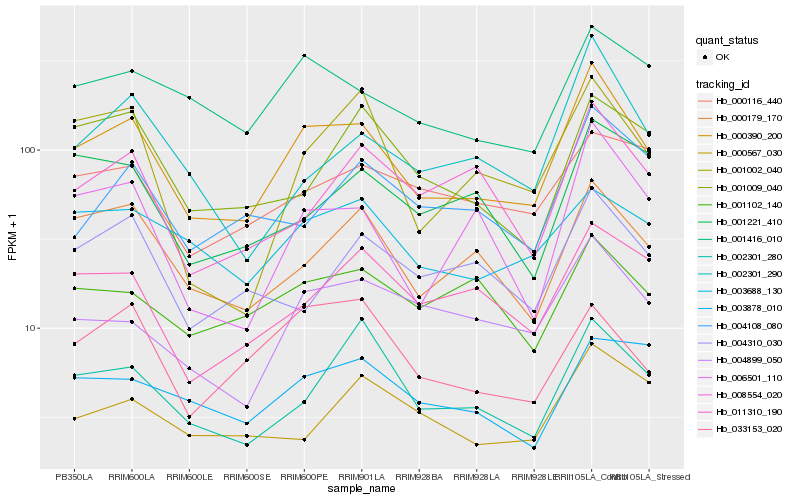
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 2 |
Hb_000179_170 |
0.1283382322 |
- |
- |
PREDICTED: protein bem46 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001416_010 |
0.1297046548 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 4 |
Hb_006501_110 |
0.1341664295 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004899_050 |
0.1365776712 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 6 |
Hb_002301_290 |
0.1401696424 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_011310_190 |
0.1418484061 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002301_280 |
0.1450912637 |
transcription factor |
TF Family: SBP |
hypothetical protein JCGZ_21462 [Jatropha curcas] |
| 9 |
Hb_003688_130 |
0.1464255133 |
- |
- |
phosphoserine phosphatase, putative [Ricinus communis] |
| 10 |
Hb_008554_020 |
0.1491180292 |
- |
- |
- |
| 11 |
Hb_001002_040 |
0.1510930573 |
- |
- |
PREDICTED: ATP-citrate synthase beta chain protein 2 [Gossypium raimondii] |
| 12 |
Hb_004310_030 |
0.1517869953 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 13 |
Hb_033153_020 |
0.1519840217 |
transcription factor |
TF Family: ARF |
Auxin response factor, putative [Ricinus communis] |
| 14 |
Hb_000116_440 |
0.1527573363 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 6-2 [Jatropha curcas] |
| 15 |
Hb_001102_140 |
0.1528422672 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 2-like [Jatropha curcas] |
| 16 |
Hb_004108_080 |
0.1536232812 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase [Vitis vinifera] |
| 17 |
Hb_001221_410 |
0.1540954829 |
- |
- |
PREDICTED: gamma carbonic anhydrase-like 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000567_030 |
0.1559370258 |
- |
- |
hypothetical protein JCGZ_21550 [Jatropha curcas] |
| 19 |
Hb_001009_040 |
0.1566148523 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Jatropha curcas] |
| 20 |
Hb_003878_010 |
0.1566682612 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |