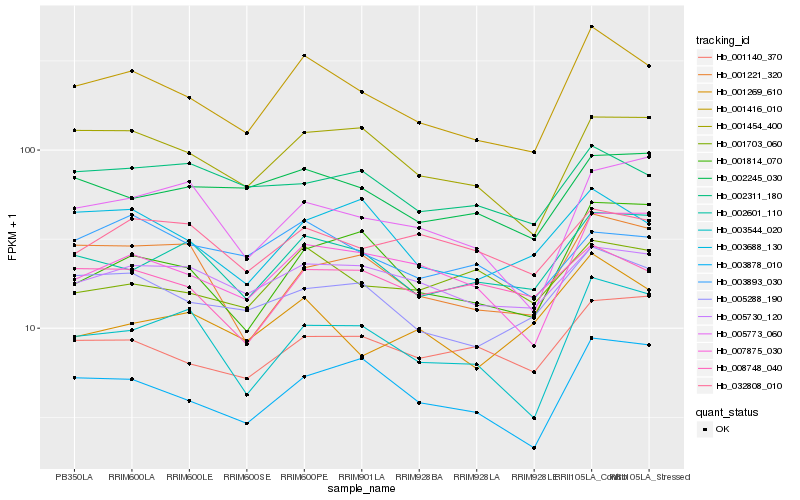
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001416_010 |
0.0 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 2 |
Hb_005288_190 |
0.0934563863 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 3 |
Hb_001454_400 |
0.0971611966 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_007875_030 |
0.1015931177 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 5 |
Hb_002601_110 |
0.1033295564 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 6 |
Hb_003878_010 |
0.1037437538 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 7 |
Hb_003544_020 |
0.1070684152 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 8 |
Hb_001221_320 |
0.1081152773 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003688_130 |
0.1099287821 |
- |
- |
phosphoserine phosphatase, putative [Ricinus communis] |
| 10 |
Hb_005730_120 |
0.1131450582 |
- |
- |
PREDICTED: protein WHI4 [Jatropha curcas] |
| 11 |
Hb_002245_030 |
0.1164867341 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_001814_070 |
0.1175767487 |
- |
- |
PREDICTED: uncharacterized protein LOC105650638 [Jatropha curcas] |
| 13 |
Hb_001140_370 |
0.1190685238 |
- |
- |
PREDICTED: uncharacterized protein LOC105648463 [Jatropha curcas] |
| 14 |
Hb_001269_610 |
0.1217297265 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_008748_040 |
0.1248653686 |
- |
- |
beta-hydroxyacyl-acyl carrier protein dehydratase [Hevea brasiliensis] |
| 16 |
Hb_005773_060 |
0.12564079 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 17 |
Hb_001703_060 |
0.1260070918 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003893_030 |
0.1260105671 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 19 |
Hb_002311_180 |
0.1266374488 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 20 |
Hb_032808_010 |
0.1271970822 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |