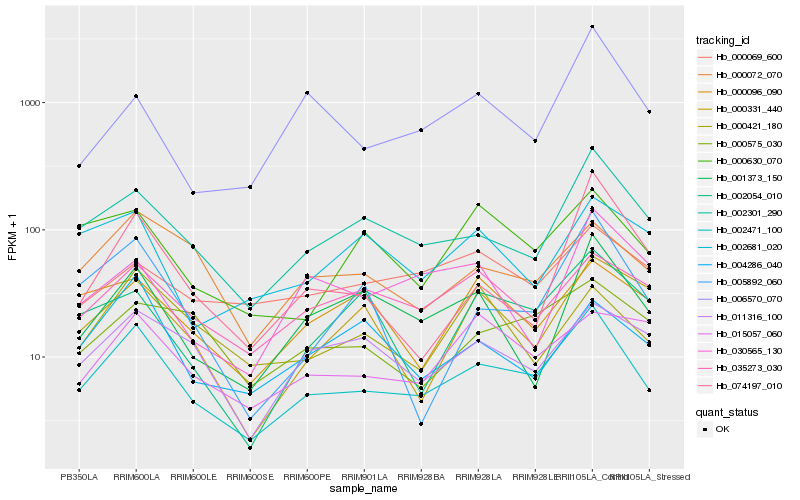
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_100 |
0.0 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002301_290 |
0.1301701138 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 3 |
Hb_001373_150 |
0.1642967833 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 4 |
Hb_000072_070 |
0.1713612924 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl diphosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_035273_030 |
0.1717459688 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 6 |
Hb_011316_100 |
0.1776526269 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4-like [Jatropha curcas] |
| 7 |
Hb_005892_060 |
0.1823406598 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000630_070 |
0.1847017482 |
- |
- |
PREDICTED: cell division protein FtsZ homolog 2-1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000575_030 |
0.1853950262 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_004286_040 |
0.1861406751 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 11 |
Hb_074197_010 |
0.1898366215 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 12 |
Hb_006570_070 |
0.189991062 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 13 |
Hb_000069_600 |
0.1926278435 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 14 |
Hb_000331_440 |
0.1933123931 |
- |
- |
hypothetical protein POPTR_0015s10290g [Populus trichocarpa] |
| 15 |
Hb_002054_010 |
0.1934986011 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 16 |
Hb_015057_060 |
0.193898246 |
- |
- |
- |
| 17 |
Hb_000096_090 |
0.194455427 |
- |
- |
hypothetical protein JCGZ_12551 [Jatropha curcas] |
| 18 |
Hb_030565_130 |
0.1946977802 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 19 |
Hb_002681_020 |
0.1949680773 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 20 |
Hb_000421_180 |
0.1950549274 |
- |
- |
ATP binding protein, putative [Ricinus communis] |