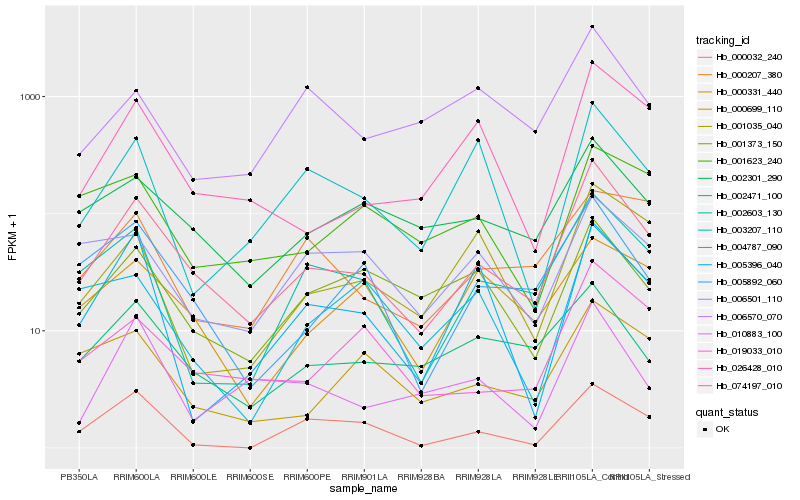
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074197_010 |
0.0 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 2 |
Hb_002603_130 |
0.1721429687 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 3 |
Hb_005892_060 |
0.1737247449 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 4 |
Hb_019033_010 |
0.1755715652 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 5 |
Hb_002301_290 |
0.1778245681 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 6 |
Hb_026428_010 |
0.1820471465 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 7 |
Hb_002471_100 |
0.1898366215 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000699_110 |
0.2011613075 |
- |
- |
hypothetical protein POPTR_0013s04180g [Populus trichocarpa] |
| 9 |
Hb_001373_150 |
0.203071002 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 10 |
Hb_010883_100 |
0.2039810351 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 11 |
Hb_000032_240 |
0.2041125457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001035_040 |
0.2094363656 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_003207_110 |
0.2164525775 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 14 |
Hb_006501_110 |
0.2174927044 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004787_090 |
0.2184310293 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 16 |
Hb_006570_070 |
0.2186192464 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 17 |
Hb_005396_040 |
0.2190371211 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001623_240 |
0.2205657456 |
- |
- |
hypothetical protein VITISV_004933 [Vitis vinifera] |
| 19 |
Hb_000207_380 |
0.221047369 |
- |
- |
sucrose transporter 6 [Hevea brasiliensis] |
| 20 |
Hb_000331_440 |
0.2210611316 |
- |
- |
hypothetical protein POPTR_0015s10290g [Populus trichocarpa] |