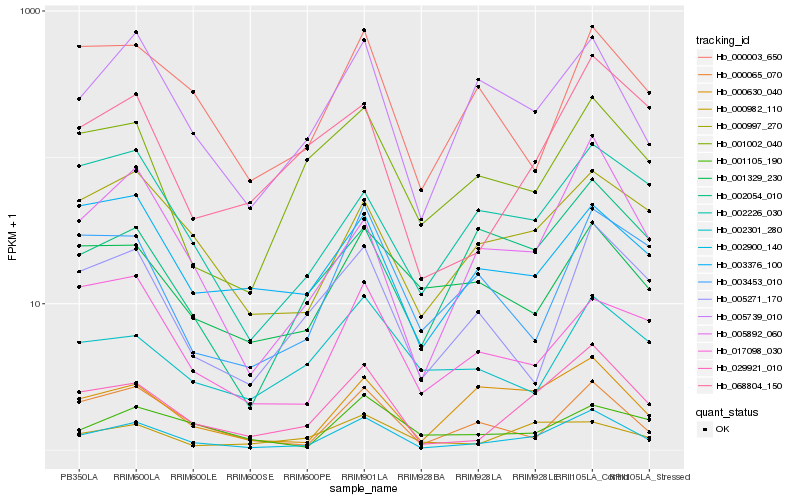
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002900_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_029921_010 |
0.104463689 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 3 |
Hb_005739_010 |
0.1538551261 |
- |
- |
hypothetical protein JCGZ_04016 [Jatropha curcas] |
| 4 |
Hb_000065_070 |
0.1631338718 |
- |
- |
hypothetical protein CISIN_1g039077mg [Citrus sinensis] |
| 5 |
Hb_005892_060 |
0.167052944 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000997_270 |
0.1719964265 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 7 |
Hb_000630_040 |
0.1762872285 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 8 |
Hb_001002_040 |
0.1834419843 |
- |
- |
PREDICTED: ATP-citrate synthase beta chain protein 2 [Gossypium raimondii] |
| 9 |
Hb_005271_170 |
0.1843972238 |
- |
- |
PREDICTED: tetraspanin-10 isoform X3 [Vitis vinifera] |
| 10 |
Hb_068804_150 |
0.1882536222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000003_650 |
0.1902981376 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_003376_100 |
0.1952660562 |
- |
- |
PREDICTED: uncharacterized protein LOC105642192 [Jatropha curcas] |
| 13 |
Hb_001105_190 |
0.1962671785 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 14 |
Hb_001329_230 |
0.1984370739 |
- |
- |
PREDICTED: peptide chain release factor 1-like, mitochondrial [Jatropha curcas] |
| 15 |
Hb_003453_010 |
0.1994355999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000982_110 |
0.1995875766 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_002226_030 |
0.1999945016 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.7 [Jatropha curcas] |
| 18 |
Hb_002301_280 |
0.2004762389 |
transcription factor |
TF Family: SBP |
hypothetical protein JCGZ_21462 [Jatropha curcas] |
| 19 |
Hb_002054_010 |
0.2005689973 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 20 |
Hb_017098_030 |
0.2044909904 |
- |
- |
hypothetical protein POPTR_0001s03560g [Populus trichocarpa] |