| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
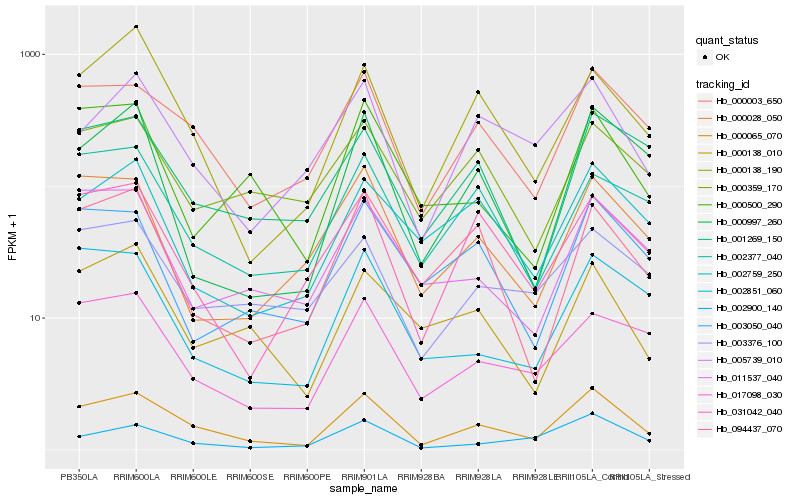
Hb_000065_070 |
0.0 |
- |
- |
hypothetical protein CISIN_1g039077mg [Citrus sinensis] |
| 2 |
Hb_000003_650 |
0.1113562238 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 3 |
Hb_000138_190 |
0.1535686686 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 4 |
Hb_017098_030 |
0.1581790016 |
- |
- |
hypothetical protein POPTR_0001s03560g [Populus trichocarpa] |
| 5 |
Hb_005739_010 |
0.1582990138 |
- |
- |
hypothetical protein JCGZ_04016 [Jatropha curcas] |
| 6 |
Hb_002900_140 |
0.1631338718 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 7 |
Hb_031042_040 |
0.1638003439 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 8 |
Hb_001269_150 |
0.1655528187 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 9 |
Hb_003376_100 |
0.1665526899 |
- |
- |
PREDICTED: uncharacterized protein LOC105642192 [Jatropha curcas] |
| 10 |
Hb_000138_010 |
0.1682078906 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 11 |
Hb_002377_040 |
0.1683649259 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_094437_070 |
0.1705678075 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 13 |
Hb_002851_060 |
0.1705823985 |
- |
- |
Clathrin light chain 2 -like protein [Gossypium arboreum] |
| 14 |
Hb_011537_040 |
0.1706596151 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 15 |
Hb_000028_050 |
0.1714104553 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 16 |
Hb_000359_170 |
0.1718346919 |
- |
- |
hypothetical protein CISIN_1g014750mg [Citrus sinensis] |
| 17 |
Hb_002759_250 |
0.1731183633 |
rubber biosynthesis |
Gene Name: Phosphomevalonate kinase |
phosphomevalonate kinase [Hevea brasiliensis] |
| 18 |
Hb_000997_260 |
0.1735479379 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003050_040 |
0.1741652287 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 20 |
Hb_000500_290 |
0.1741925122 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |