| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
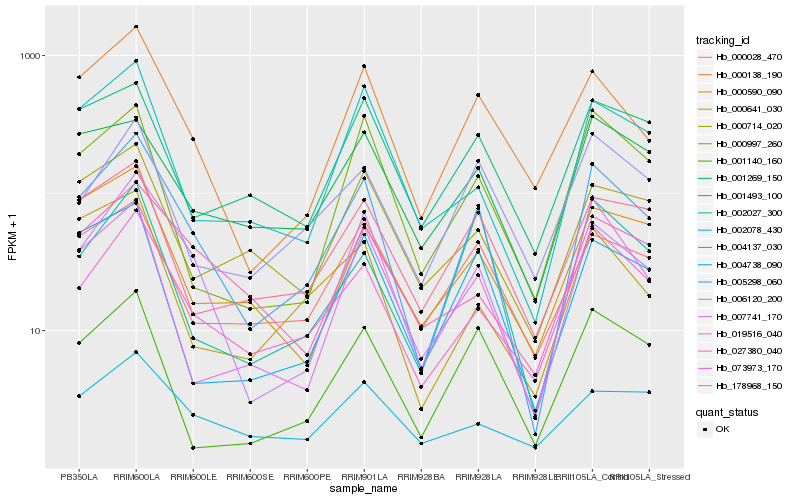
Hb_005298_060 |
0.0 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Malus domestica] |
| 2 |
Hb_007741_170 |
0.1030154697 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4-like [Jatropha curcas] |
| 3 |
Hb_027380_040 |
0.1285457987 |
- |
- |
hypothetical protein B456_011G046500, partial [Gossypium raimondii] |
| 4 |
Hb_000138_190 |
0.1302411175 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 5 |
Hb_004738_090 |
0.1601967598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_178968_150 |
0.1615956425 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_001140_160 |
0.1622764623 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_002078_430 |
0.1634075346 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 9 |
Hb_006120_200 |
0.1640898128 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 10 |
Hb_000028_470 |
0.1641249071 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 11 |
Hb_073973_170 |
0.1657790714 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 12 |
Hb_001269_150 |
0.1668138695 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 13 |
Hb_002027_300 |
0.1671781489 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 14 |
Hb_000590_090 |
0.1678136264 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_019516_040 |
0.1692068135 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 16 |
Hb_000997_260 |
0.1697730067 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001493_100 |
0.1704384722 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 34-like [Jatropha curcas] |
| 18 |
Hb_000714_020 |
0.172335366 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 19 |
Hb_004137_030 |
0.1740255211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000641_030 |
0.1747005757 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |