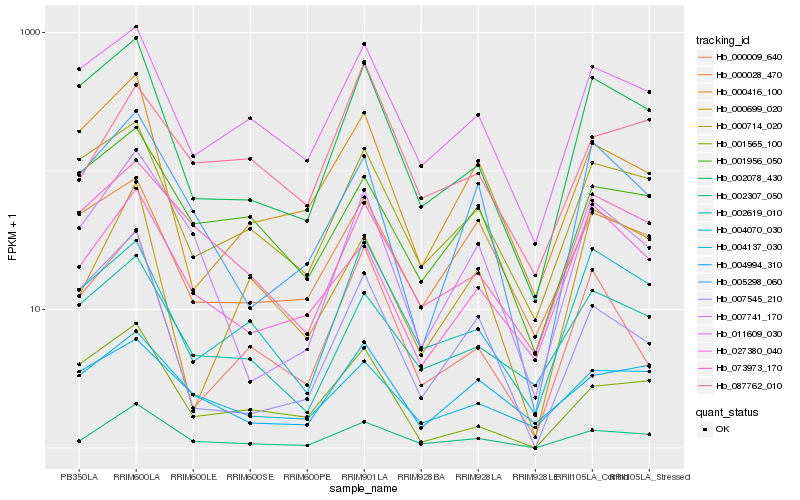
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_050 |
0.0 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007741_170 |
0.1582003764 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4-like [Jatropha curcas] |
| 3 |
Hb_002078_430 |
0.1745793927 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 4 |
Hb_005298_060 |
0.1814185163 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Malus domestica] |
| 5 |
Hb_004137_030 |
0.185353925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011609_030 |
0.1857925265 |
- |
- |
conserved hypothetical protein 13 [Hevea brasiliensis] |
| 7 |
Hb_002619_010 |
0.1887705774 |
- |
- |
- |
| 8 |
Hb_000714_020 |
0.1903725555 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 9 |
Hb_007545_210 |
0.1927243846 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |
| 10 |
Hb_000416_100 |
0.1929793586 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 11 |
Hb_000699_020 |
0.1940352158 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 12 |
Hb_027380_040 |
0.1945051637 |
- |
- |
hypothetical protein B456_011G046500, partial [Gossypium raimondii] |
| 13 |
Hb_073973_170 |
0.1961750994 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 14 |
Hb_004070_030 |
0.198058902 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_001956_050 |
0.1994520939 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 16 |
Hb_001565_100 |
0.2046627681 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000009_640 |
0.2049629247 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_12318 [Jatropha curcas] |
| 18 |
Hb_087762_010 |
0.2109220736 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 19 |
Hb_000028_470 |
0.2134860371 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004994_310 |
0.2143933235 |
- |
- |
Kiwellin, putative [Ricinus communis] |