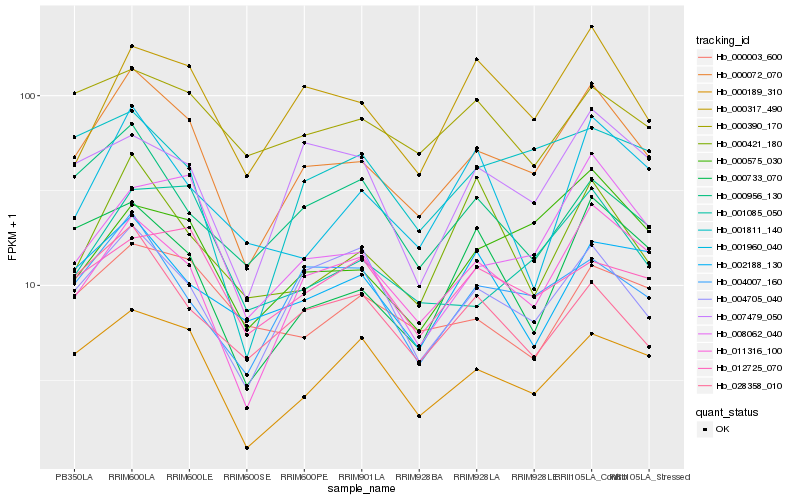
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_070 |
0.0 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_011316_100 |
0.1041412916 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4-like [Jatropha curcas] |
| 3 |
Hb_000189_310 |
0.1223968975 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004007_160 |
0.1318340262 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 5 |
Hb_008062_040 |
0.1318367537 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001085_050 |
0.1329911007 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 7 |
Hb_000575_030 |
0.1330914825 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000317_490 |
0.1335775104 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 9 |
Hb_004705_040 |
0.1339305292 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 10 |
Hb_000733_070 |
0.1414418418 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 11 |
Hb_007479_050 |
0.1458263539 |
- |
- |
PREDICTED: uncharacterized protein LOC105649412 [Jatropha curcas] |
| 12 |
Hb_001811_140 |
0.146277466 |
- |
- |
hypothetical protein POPTR_0007s03140g [Populus trichocarpa] |
| 13 |
Hb_000003_600 |
0.1478932274 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 14 |
Hb_000956_130 |
0.1483618128 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 15 |
Hb_000390_170 |
0.1490659683 |
- |
- |
PREDICTED: V-type proton ATPase subunit H [Jatropha curcas] |
| 16 |
Hb_028358_010 |
0.1499837516 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002188_130 |
0.1511030589 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000421_180 |
0.1535126494 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_001960_040 |
0.1543838865 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 20 |
Hb_012725_070 |
0.1549896938 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |