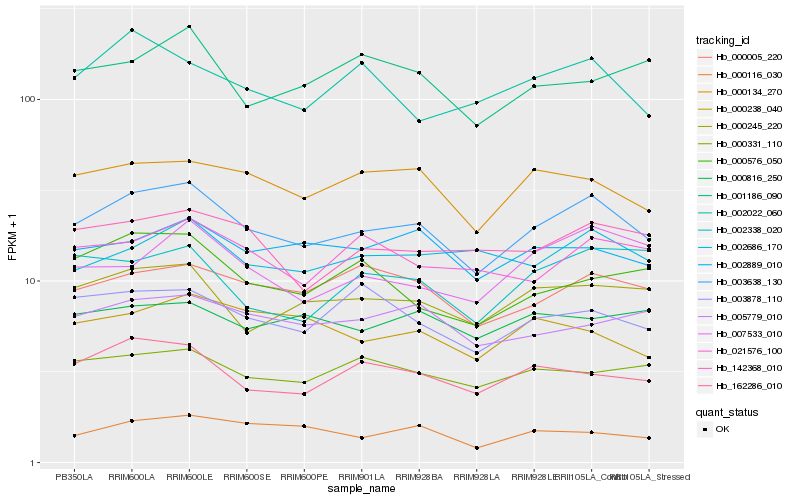
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003638_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645779 [Jatropha curcas] |
| 2 |
Hb_000245_220 |
0.0856073609 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 3 |
Hb_000005_220 |
0.0892888054 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 4 |
Hb_000134_270 |
0.0893539698 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 5 |
Hb_162286_010 |
0.0904475792 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 6 |
Hb_002889_010 |
0.094937121 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 7 |
Hb_142368_010 |
0.0961673618 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000331_110 |
0.0985711141 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 9 |
Hb_002686_170 |
0.0989936038 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005779_010 |
0.0997594709 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_003878_110 |
0.1000249327 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 12 |
Hb_021576_100 |
0.1004269001 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 13 |
Hb_007533_010 |
0.103511466 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 14 |
Hb_000116_030 |
0.1042638477 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 15 |
Hb_000238_040 |
0.1068818257 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 16 |
Hb_002022_060 |
0.1075159986 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 17 |
Hb_000576_050 |
0.1076450539 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 18 |
Hb_000816_250 |
0.107936145 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002338_020 |
0.1084546377 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 20 |
Hb_001186_090 |
0.1087608914 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |