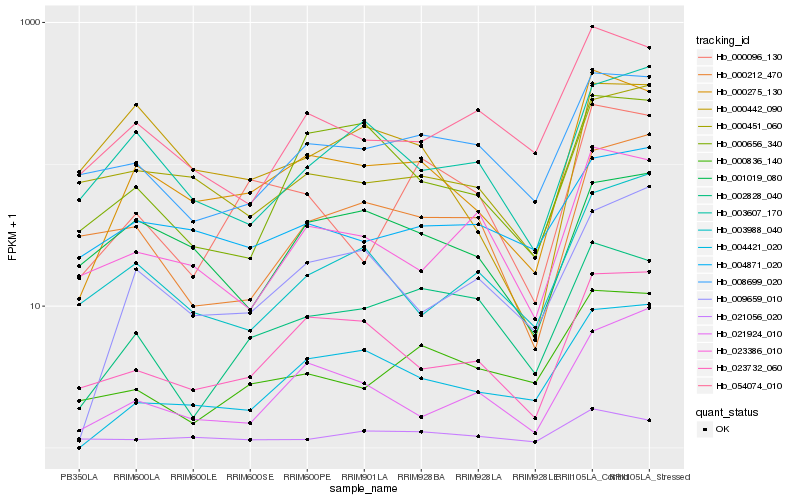
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000275_130 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |
| 2 |
Hb_023732_060 |
0.1522939344 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 3 |
Hb_054074_010 |
0.1640768206 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 4 |
Hb_004421_020 |
0.1642208126 |
- |
- |
- |
| 5 |
Hb_008699_020 |
0.1727459786 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 6 |
Hb_000096_130 |
0.174601316 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 7 |
Hb_023386_010 |
0.1761160944 |
- |
- |
PREDICTED: histidine triad nucleotide-binding protein 3 [Jatropha curcas] |
| 8 |
Hb_000451_060 |
0.1783971517 |
- |
- |
hypothetical protein CICLE_v10010074mg [Citrus clementina] |
| 9 |
Hb_009659_010 |
0.1792263539 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Vitis vinifera] |
| 10 |
Hb_000836_140 |
0.1832100076 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002828_040 |
0.1833738679 |
- |
- |
transmembrane protein, putative [Medicago truncatula] |
| 12 |
Hb_003607_170 |
0.1853439356 |
- |
- |
U6 snRNA-associated Sm-like protein LSm5 [Hevea brasiliensis] |
| 13 |
Hb_000656_340 |
0.1859978883 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 14 |
Hb_021056_020 |
0.186658422 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 15 |
Hb_004871_020 |
0.1899045405 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 16 |
Hb_021924_010 |
0.192917704 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 17 |
Hb_003988_040 |
0.193313993 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 18 |
Hb_000212_470 |
0.2019420056 |
- |
- |
hypothetical protein CICLE_v10033256mg [Citrus clementina] |
| 19 |
Hb_001019_080 |
0.202339302 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 20 |
Hb_000442_090 |
0.2051887763 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |