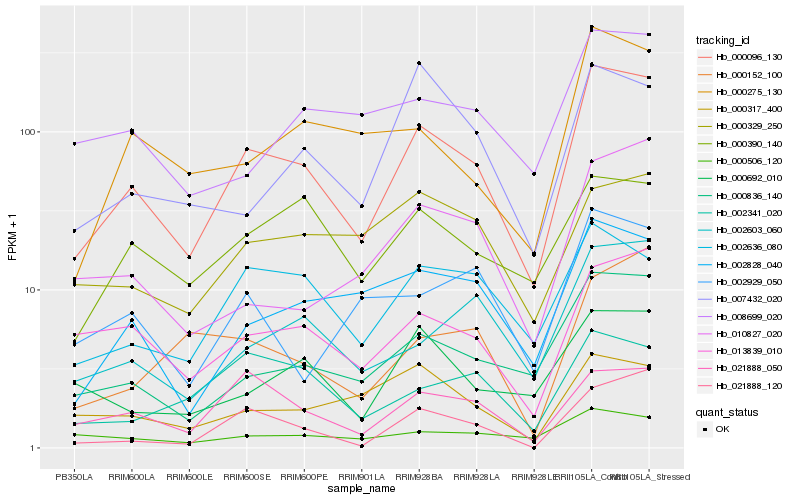
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000096_130 |
0.0 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 2 |
Hb_000836_140 |
0.1364976101 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 3 |
Hb_013839_010 |
0.1572504333 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 4 |
Hb_002603_060 |
0.1631983949 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 5 |
Hb_002828_040 |
0.1666215906 |
- |
- |
transmembrane protein, putative [Medicago truncatula] |
| 6 |
Hb_002636_080 |
0.170959143 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 7 |
Hb_021888_050 |
0.171614635 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 8 |
Hb_021888_120 |
0.174108922 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 9 |
Hb_000275_130 |
0.174601316 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |
| 10 |
Hb_000692_010 |
0.184340776 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |
| 11 |
Hb_008699_020 |
0.1849946989 |
- |
- |
eukaryotic translation initiation factor 5A isoform IV [Hevea brasiliensis] |
| 12 |
Hb_000390_140 |
0.1852806748 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 13 |
Hb_010827_020 |
0.1870286042 |
- |
- |
60S ribosomal protein L17B [Hevea brasiliensis] |
| 14 |
Hb_000506_120 |
0.1873396588 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 15 |
Hb_002929_050 |
0.1913432936 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000317_400 |
0.1940547825 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002341_020 |
0.1944955085 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 18 |
Hb_007432_020 |
0.1952786361 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 19 |
Hb_000329_250 |
0.1968491752 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000152_100 |
0.1975278678 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |