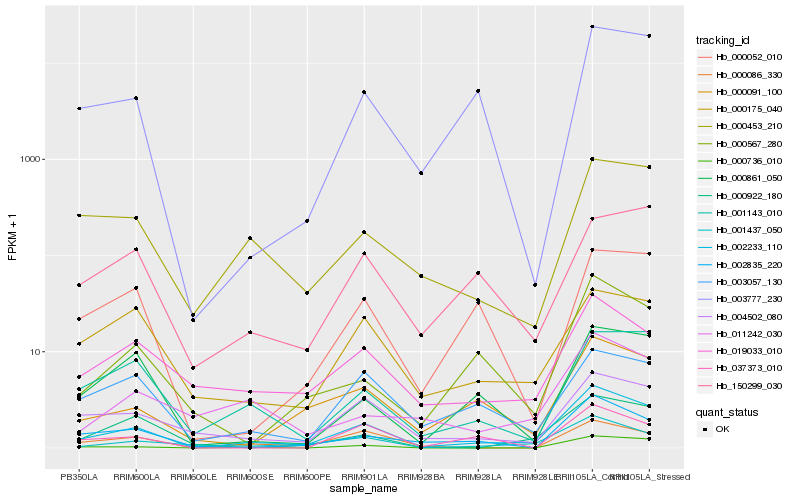
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002233_110 |
0.0 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 2 |
Hb_001437_050 |
0.1784462758 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 3 |
Hb_019033_010 |
0.194742179 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 4 |
Hb_000091_100 |
0.1950397644 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_004502_080 |
0.1953369623 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 6 |
Hb_000736_010 |
0.2009624661 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 7 |
Hb_002835_220 |
0.2042548312 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 8 |
Hb_000453_210 |
0.2050803837 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 9 |
Hb_001143_010 |
0.2066901257 |
- |
- |
- |
| 10 |
Hb_000567_280 |
0.210475721 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 11 |
Hb_003777_230 |
0.2120492593 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 12 |
Hb_003057_130 |
0.216907081 |
- |
- |
- |
| 13 |
Hb_000086_330 |
0.2175761298 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000922_180 |
0.2218574651 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 15 |
Hb_000175_040 |
0.2223292126 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 16 |
Hb_000861_050 |
0.2246878309 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000052_010 |
0.2253648954 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 18 |
Hb_037373_010 |
0.2277899874 |
- |
- |
PREDICTED: uncharacterized protein LOC102621950 [Citrus sinensis] |
| 19 |
Hb_011242_030 |
0.2289019798 |
- |
- |
- |
| 20 |
Hb_150299_030 |
0.2296033386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |