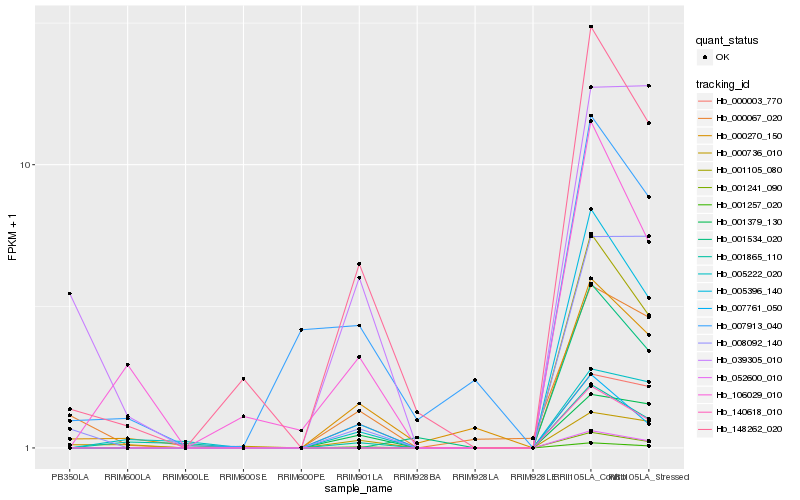
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148262_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001257_020 |
0.1492417589 |
- |
- |
- |
| 3 |
Hb_106029_010 |
0.1749109301 |
- |
- |
Transcription factor ICE1, putative [Ricinus communis] |
| 4 |
Hb_000270_150 |
0.1764686923 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001379_130 |
0.1787797354 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 6 |
Hb_000736_010 |
0.1813407244 |
- |
- |
PREDICTED: uncharacterized protein LOC105762083 [Gossypium raimondii] |
| 7 |
Hb_000003_770 |
0.1883403621 |
- |
- |
- |
| 8 |
Hb_007761_050 |
0.201171526 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 9 |
Hb_000067_020 |
0.2192273918 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 10 |
Hb_001865_110 |
0.2258167354 |
- |
- |
- |
| 11 |
Hb_005222_020 |
0.2317800841 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_001534_020 |
0.2328267126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_039305_010 |
0.2341840113 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 14 |
Hb_008092_140 |
0.236063924 |
- |
- |
PREDICTED: uncharacterized protein LOC103636948 [Zea mays] |
| 15 |
Hb_001105_080 |
0.2362790879 |
- |
- |
- |
| 16 |
Hb_005396_140 |
0.2366258825 |
- |
- |
- |
| 17 |
Hb_140618_010 |
0.2368169681 |
- |
- |
- |
| 18 |
Hb_001241_090 |
0.2368195978 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 19 |
Hb_052600_010 |
0.23682943 |
- |
- |
- |
| 20 |
Hb_007913_040 |
0.237431265 |
- |
- |
sterol desaturase, putative [Ricinus communis] |