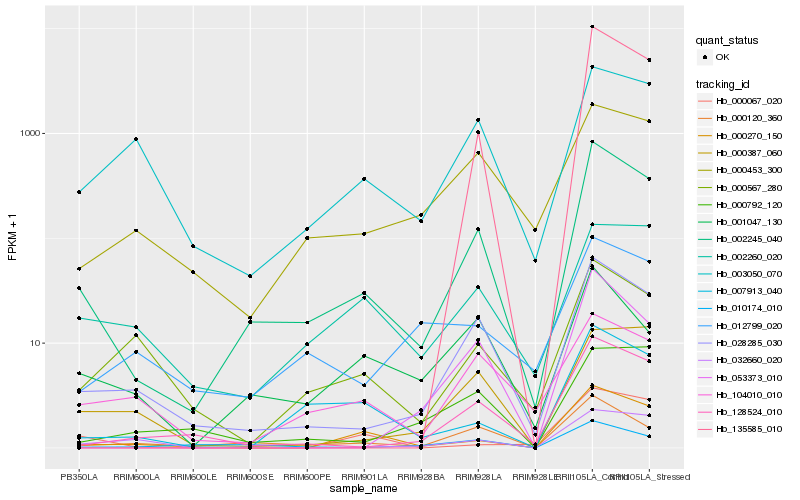
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002245_040 |
0.0 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 2 |
Hb_028285_030 |
0.1418735222 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 3 |
Hb_128524_010 |
0.1692086568 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000120_360 |
0.1954727263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000270_150 |
0.1985750002 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_135585_010 |
0.2002617287 |
- |
- |
- |
| 7 |
Hb_007913_040 |
0.2110141982 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 8 |
Hb_053373_010 |
0.2122471329 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 9 |
Hb_000453_300 |
0.2272406294 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 10 |
Hb_032660_020 |
0.2282865957 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 11 |
Hb_000567_280 |
0.2317869935 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 12 |
Hb_010174_010 |
0.2327723035 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 13 |
Hb_000387_060 |
0.2369770757 |
- |
- |
- |
| 14 |
Hb_003050_070 |
0.2391667658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012799_020 |
0.2398842312 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_104010_010 |
0.2414465917 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |
| 17 |
Hb_001047_130 |
0.2424765371 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 18 |
Hb_002260_020 |
0.2435784136 |
- |
- |
- |
| 19 |
Hb_000067_020 |
0.2460029883 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 20 |
Hb_000792_120 |
0.2460030236 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |