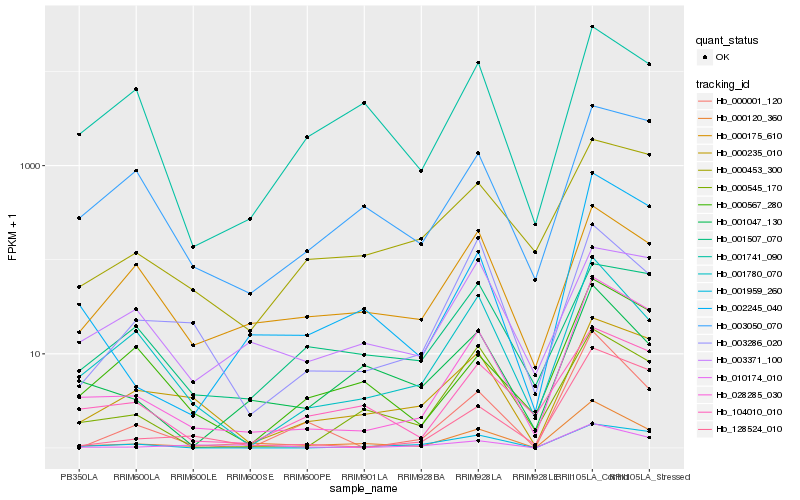
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_360 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001047_130 |
0.1571266089 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 3 |
Hb_010174_010 |
0.1629166889 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 4 |
Hb_028285_030 |
0.1750498381 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 5 |
Hb_000175_610 |
0.1893453649 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 6 |
Hb_002245_040 |
0.1954727263 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 7 |
Hb_001780_070 |
0.1982542382 |
- |
- |
hypothetical protein JCGZ_08549 [Jatropha curcas] |
| 8 |
Hb_001741_090 |
0.209736372 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 9 |
Hb_000235_010 |
0.2136032915 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 10 |
Hb_000001_120 |
0.2157917096 |
- |
- |
hypothetical protein JCGZ_03063 [Jatropha curcas] |
| 11 |
Hb_104010_010 |
0.2180096212 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |
| 12 |
Hb_003050_070 |
0.2265995965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000567_280 |
0.2278600564 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 14 |
Hb_003286_020 |
0.2293766449 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 15 |
Hb_000545_170 |
0.2323580119 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 11, plasma membrane-type isoform X2 [Jatropha curcas] |
| 16 |
Hb_000453_300 |
0.2332652348 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 17 |
Hb_003371_100 |
0.2373534626 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_128524_010 |
0.2400989875 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001507_070 |
0.2492913813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001959_260 |
0.2525485575 |
- |
- |
- |