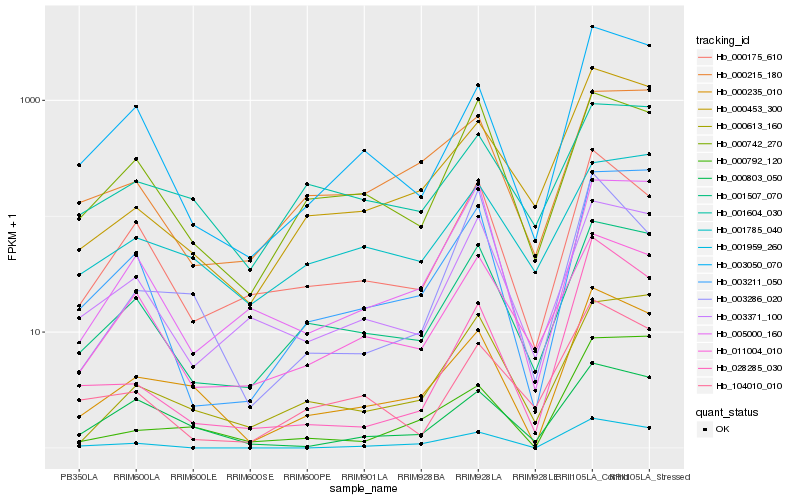
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000235_010 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 2 |
Hb_003050_070 |
0.1462185879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000803_050 |
0.1496836742 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 4 |
Hb_001507_070 |
0.1559022318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000453_300 |
0.1596609456 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 6 |
Hb_003211_050 |
0.1665691931 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000175_610 |
0.1696333252 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000792_120 |
0.1779435283 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000613_160 |
0.1783389597 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 10 |
Hb_000742_270 |
0.1788556469 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 11 |
Hb_001959_260 |
0.1815633273 |
- |
- |
- |
| 12 |
Hb_011004_010 |
0.1824343854 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 13 |
Hb_003371_100 |
0.1842696364 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 14 |
Hb_001604_030 |
0.1850379691 |
- |
- |
ubiquitin-conjugating enzyme E2 35-like [Cucumis sativus] |
| 15 |
Hb_005000_160 |
0.1872060445 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 16 |
Hb_000215_180 |
0.1901011418 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 17 |
Hb_003286_020 |
0.1911417279 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 18 |
Hb_028285_030 |
0.1944493197 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 19 |
Hb_001785_040 |
0.1956785173 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 20 |
Hb_104010_010 |
0.1968433419 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |