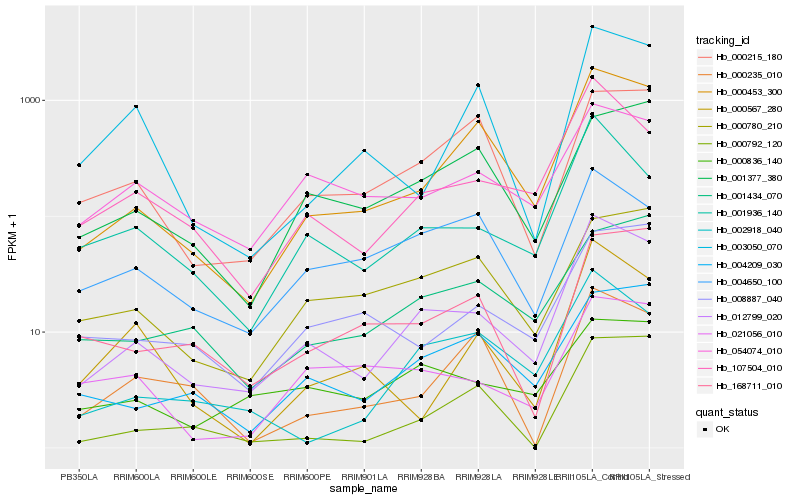
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012799_020 |
0.0 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_000453_300 |
0.1407149438 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 3 |
Hb_107504_010 |
0.1429769731 |
- |
- |
- |
| 4 |
Hb_001936_140 |
0.1599515372 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 1 [Jatropha curcas] |
| 5 |
Hb_002918_040 |
0.1814229246 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 6 |
Hb_004209_030 |
0.1854062309 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000792_120 |
0.2014436237 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_168711_010 |
0.2014871265 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 [Sesamum indicum] |
| 9 |
Hb_000836_140 |
0.2017605618 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 10 |
Hb_021056_010 |
0.2038760623 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_008887_040 |
0.2056269725 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_001434_070 |
0.2057958506 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 13 |
Hb_054074_010 |
0.2058873789 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 14 |
Hb_000567_280 |
0.2081604452 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 15 |
Hb_003050_070 |
0.2125153269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000780_210 |
0.2126841538 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 17 |
Hb_000235_010 |
0.2148839399 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 18 |
Hb_000215_180 |
0.2150969909 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 19 |
Hb_004650_100 |
0.2153071392 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001377_380 |
0.2159151857 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |