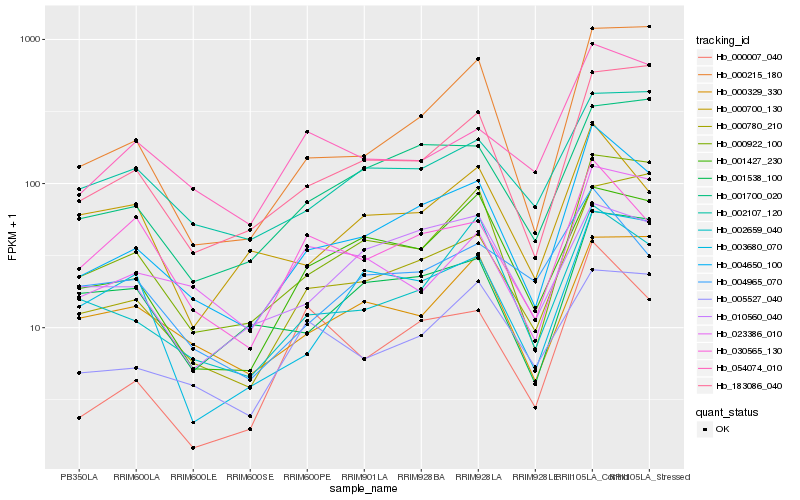
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004650_100 |
0.0 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000922_100 |
0.1264067727 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 3 |
Hb_002659_040 |
0.1351420668 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_001700_020 |
0.1523550903 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 5 |
Hb_001427_230 |
0.1525605394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_183086_040 |
0.1528283708 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 7 |
Hb_000007_040 |
0.1530905958 |
- |
- |
hypothetical protein MIMGU_mgv1a017616mg [Erythranthe guttata] |
| 8 |
Hb_000215_180 |
0.1552520604 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 9 |
Hb_001538_100 |
0.1590323739 |
- |
- |
PREDICTED: histone deacetylase 6 [Jatropha curcas] |
| 10 |
Hb_030565_130 |
0.1597343761 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 11 |
Hb_054074_010 |
0.1598258867 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 12 |
Hb_004965_070 |
0.1600941583 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 13 |
Hb_000700_130 |
0.1611683821 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005527_040 |
0.163244531 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 15 |
Hb_002107_120 |
0.1633768222 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 16 |
Hb_023386_010 |
0.1637553774 |
- |
- |
PREDICTED: histidine triad nucleotide-binding protein 3 [Jatropha curcas] |
| 17 |
Hb_000329_330 |
0.163798843 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 18 |
Hb_000780_210 |
0.1665551783 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 19 |
Hb_003680_070 |
0.1666221237 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010560_040 |
0.1670528167 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |