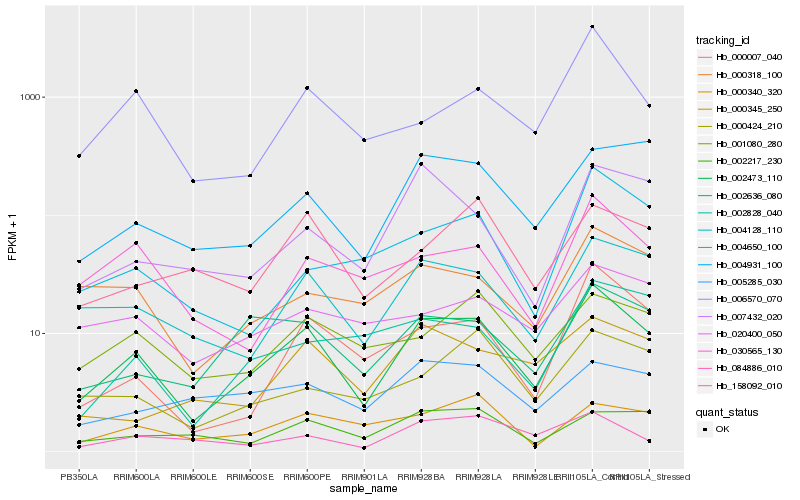
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002473_110 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_004128_110 |
0.1516830482 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 3 |
Hb_000007_040 |
0.175059069 |
- |
- |
hypothetical protein MIMGU_mgv1a017616mg [Erythranthe guttata] |
| 4 |
Hb_002636_080 |
0.1754673326 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 5 |
Hb_006570_070 |
0.181642318 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 6 |
Hb_004931_100 |
0.1851597081 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000318_100 |
0.1876496145 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 8 |
Hb_030565_130 |
0.1882624245 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 9 |
Hb_001080_280 |
0.1924477766 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 10 |
Hb_000424_210 |
0.1979792003 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002828_040 |
0.1988789228 |
- |
- |
transmembrane protein, putative [Medicago truncatula] |
| 12 |
Hb_084886_010 |
0.1995150066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002217_230 |
0.201087183 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 14 |
Hb_007432_020 |
0.201306821 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 15 |
Hb_000340_320 |
0.2018456368 |
- |
- |
PREDICTED: uncharacterized protein LOC105650052 [Jatropha curcas] |
| 16 |
Hb_000345_250 |
0.2022697342 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 17 |
Hb_158092_010 |
0.2032222379 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 18 |
Hb_004650_100 |
0.2042993827 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_020400_050 |
0.2053413593 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 20 |
Hb_005285_030 |
0.2088160382 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |