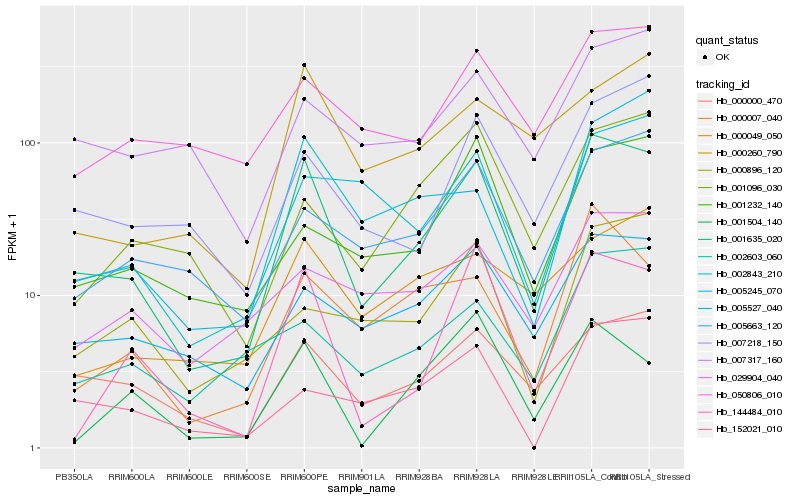
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_020 |
0.0 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 2 |
Hb_000000_470 |
0.1663371919 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X4 [Jatropha curcas] |
| 3 |
Hb_005527_040 |
0.1731250086 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 4 |
Hb_144484_010 |
0.1765254883 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000007_040 |
0.1832139482 |
- |
- |
hypothetical protein MIMGU_mgv1a017616mg [Erythranthe guttata] |
| 6 |
Hb_005663_120 |
0.1912398149 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 7 |
Hb_007218_150 |
0.1929186114 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 8 |
Hb_050806_010 |
0.1973458947 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 9 |
Hb_002843_210 |
0.1982427287 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 10 |
Hb_029904_040 |
0.1983166085 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 11 |
Hb_002603_060 |
0.1997422868 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 12 |
Hb_001232_140 |
0.2008084026 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 13 |
Hb_005245_070 |
0.2012922943 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 14 |
Hb_001504_140 |
0.2049581375 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 15 |
Hb_000260_790 |
0.2068583492 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 16 |
Hb_007317_160 |
0.2078330057 |
- |
- |
PREDICTED: uncharacterized protein At2g27730, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000049_050 |
0.2085170519 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 18 |
Hb_152021_010 |
0.2086059445 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000896_120 |
0.2089010002 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 20 |
Hb_001096_030 |
0.2089028209 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |