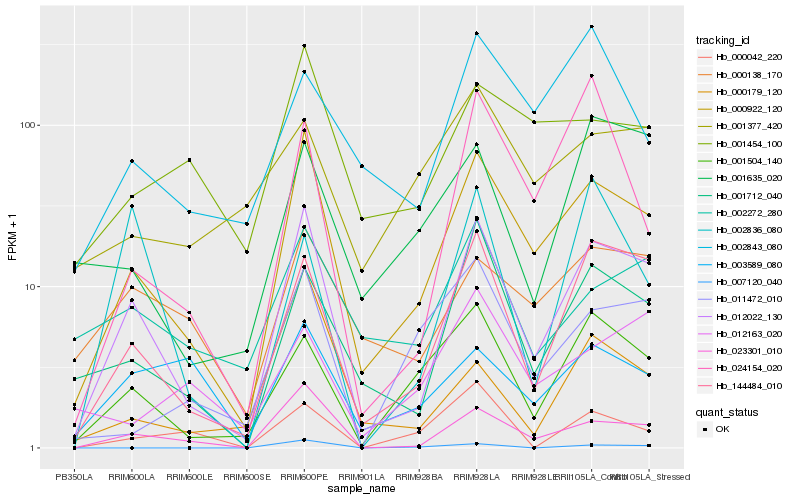
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012022_130 |
0.0 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 2 |
Hb_000922_120 |
0.118233615 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 3 |
Hb_023301_010 |
0.141912053 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 4 |
Hb_144484_010 |
0.1465427699 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_001712_040 |
0.2040800602 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 6 |
Hb_001504_140 |
0.2229750266 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 7 |
Hb_000138_170 |
0.2546771468 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 8 |
Hb_002272_280 |
0.2550150624 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 9 |
Hb_001635_020 |
0.2582298277 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 10 |
Hb_011472_010 |
0.2620174948 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 11 |
Hb_024154_020 |
0.2630790053 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 12 |
Hb_001454_100 |
0.2732897809 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 13 |
Hb_000179_120 |
0.273974654 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 14 |
Hb_003589_080 |
0.2765147946 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 15 |
Hb_002836_080 |
0.2773932116 |
- |
- |
- |
| 16 |
Hb_000042_220 |
0.2787025053 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |
| 17 |
Hb_007120_040 |
0.2811025291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002843_080 |
0.2815362831 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 19 |
Hb_012163_020 |
0.294444404 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 20 |
Hb_001377_420 |
0.2971627019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |